Introduction and setup
Overview
Teaching: XX min
Exercises: XX minQuestions
Install required packages.
Download the data.
Download data
The data we will use in this lesson is obtained from the Gene Expression Omnibus, accession number GSE96870.
dir.create("data", showWarnings = FALSE)
download.file(
url = "https://github.com/Bioconductor/bioconductor-teaching/blob/master/data/GSE96870/GSE96870_counts_cerebellum.csv?raw=true",
destfile = "data/GSE96870_counts_cerebellum.csv"
)
download.file(
url = "https://github.com/Bioconductor/bioconductor-teaching/blob/master/data/GSE96870/GSE96870_coldata_cerebellum.csv?raw=true",
destfile = "data/GSE96870_coldata_cerebellum.csv"
)
download.file(
url = "https://github.com/Bioconductor/bioconductor-teaching/blob/master/data/GSE96870/GSE96870_coldata_all.csv?raw=true",
destfile = "data/GSE96870_coldata_all.csv"
)
download.file(
url = "https://github.com/Bioconductor/bioconductor-teaching/blob/master/data/GSE96870/GSE96870_rowranges.tsv?raw=true",
destfile = "data/GSE96870_rowranges.tsv"
)
Key Points
Key point 1
Introduction to RNA-seq
Overview
Teaching: XX min
Exercises: XX minQuestions
What is RNA-seq?
Objectives
Explain what RNA-seq is and what the generated data looks like.
Provide an overview of common quality control steps for the raw data.
Explain how gene expression levels can be estimated from raw data.
Contribute!
This episode is intended to introduce important concepts in RNA-seq data, to bring everyone up to speed.
Key Points
Key point 1
Experimental design
Overview
Teaching: XX min
Exercises: XX minQuestions
How do we design experiments optimally?
How do we interpret a given design?
Objectives
Explain the formula notation and design matrices.
Explore different designs and learn how to interpret coefficients.
Contribute!
This episode is intended to discuss experimental design - what it means, why it is important, how you would translate your metadata into a suitable design matrix, how coefficients are to be interpreted.
suppressPackageStartupMessages({
library(SummarizedExperiment)
library(ExploreModelMatrix)
library(dplyr)
})
meta <- read.csv("data/GSE96870_coldata_all.csv", row.names = 1)
meta
title geo_accession organism age sex infection strain time tissue mouse
GSM2545336 CNS_RNA-seq_10C GSM2545336 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day8 Cerebellum 14
GSM2545337 CNS_RNA-seq_11C GSM2545337 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 9
GSM2545338 CNS_RNA-seq_12C GSM2545338 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 10
GSM2545339 CNS_RNA-seq_13C GSM2545339 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 15
GSM2545340 CNS_RNA-seq_14C GSM2545340 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Cerebellum 18
GSM2545341 CNS_RNA-seq_17C GSM2545341 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Cerebellum 6
GSM2545342 CNS_RNA-seq_1C GSM2545342 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day8 Cerebellum 5
GSM2545343 CNS_RNA-seq_20C GSM2545343 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 11
GSM2545344 CNS_RNA-seq_21C GSM2545344 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 22
GSM2545345 CNS_RNA-seq_22C GSM2545345 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Cerebellum 13
GSM2545346 CNS_RNA-seq_25C GSM2545346 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Cerebellum 23
GSM2545347 CNS_RNA-seq_26C GSM2545347 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Cerebellum 24
GSM2545348 CNS_RNA-seq_27C GSM2545348 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 8
GSM2545349 CNS_RNA-seq_28C GSM2545349 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 7
GSM2545350 CNS_RNA-seq_29C GSM2545350 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Cerebellum 1
GSM2545351 CNS_RNA-seq_2C GSM2545351 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day8 Cerebellum 16
GSM2545352 CNS_RNA-seq_30C GSM2545352 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 21
GSM2545353 CNS_RNA-seq_3C GSM2545353 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 4
GSM2545354 CNS_RNA-seq_4C GSM2545354 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 2
GSM2545355 CNS_RNA-seq_571 GSM2545355 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 1
GSM2545356 CNS_RNA-seq_574 GSM2545356 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 2
GSM2545357 CNS_RNA-seq_575 GSM2545357 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 3
GSM2545358 CNS_RNA-seq_583 GSM2545358 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 4
GSM2545359 CNS_RNA-seq_585 GSM2545359 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day8 Spinalcord 5
GSM2545360 CNS_RNA-seq_589 GSM2545360 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 6
GSM2545361 CNS_RNA-seq_590 GSM2545361 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 7
GSM2545362 CNS_RNA-seq_5C GSM2545362 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 20
GSM2545363 CNS_RNA-seq_6C GSM2545363 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Cerebellum 12
GSM2545364 CNS_RNA-seq_709 GSM2545364 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 8
GSM2545365 CNS_RNA-seq_710 GSM2545365 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 9
GSM2545366 CNS_RNA-seq_711 GSM2545366 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 10
GSM2545367 CNS_RNA-seq_713 GSM2545367 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 11
GSM2545368 CNS_RNA-seq_728 GSM2545368 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 12
GSM2545369 CNS_RNA-seq_729 GSM2545369 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 13
GSM2545370 CNS_RNA-seq_730 GSM2545370 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day8 Spinalcord 14
GSM2545371 CNS_RNA-seq_731 GSM2545371 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 15
GSM2545372 CNS_RNA-seq_733 GSM2545372 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 17
GSM2545373 CNS_RNA-seq_735 GSM2545373 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 18
GSM2545374 CNS_RNA-seq_736 GSM2545374 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day8 Spinalcord 19
GSM2545375 CNS_RNA-seq_738 GSM2545375 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 20
GSM2545376 CNS_RNA-seq_740 GSM2545376 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 21
GSM2545377 CNS_RNA-seq_741 GSM2545377 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 22
GSM2545378 CNS_RNA-seq_742 GSM2545378 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 23
GSM2545379 CNS_RNA-seq_743 GSM2545379 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 24
GSM2545380 CNS_RNA-seq_9C GSM2545380 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day8 Cerebellum 19
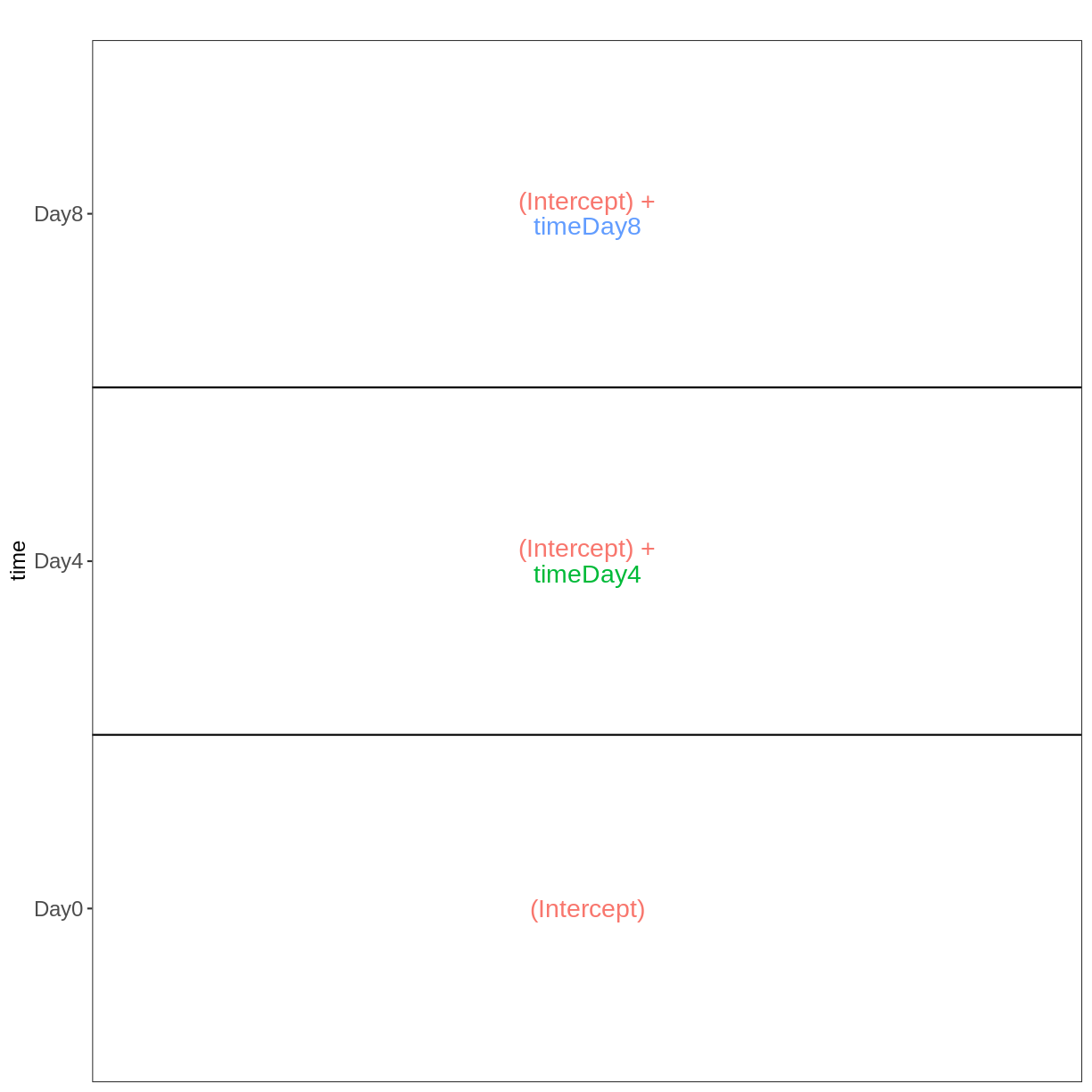
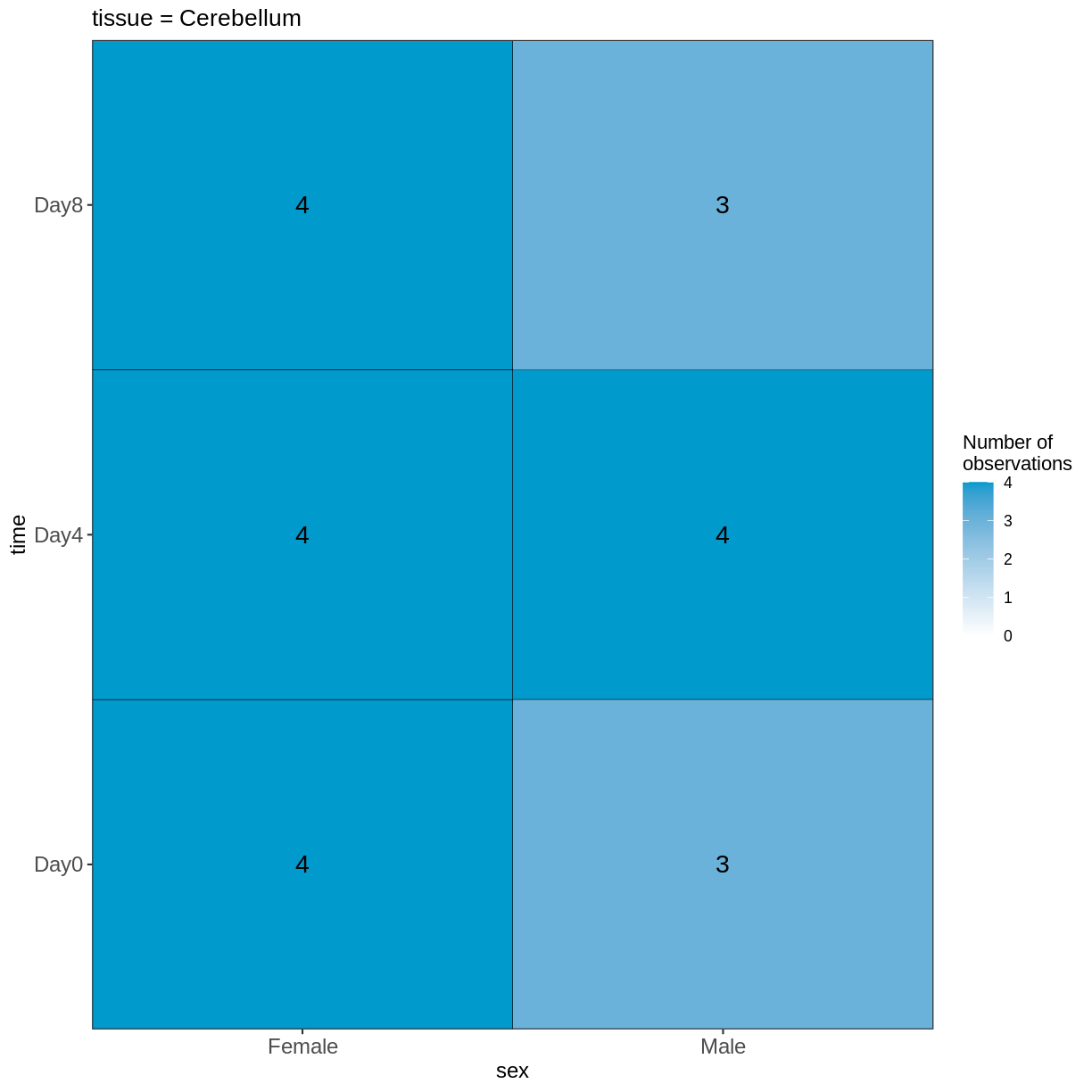
vd <- VisualizeDesign(sampleData = meta,
designFormula = ~ tissue + time + sex)
vd$cooccurrenceplots
$`tissue = Cerebellum`
$`tissue = Spinalcord`
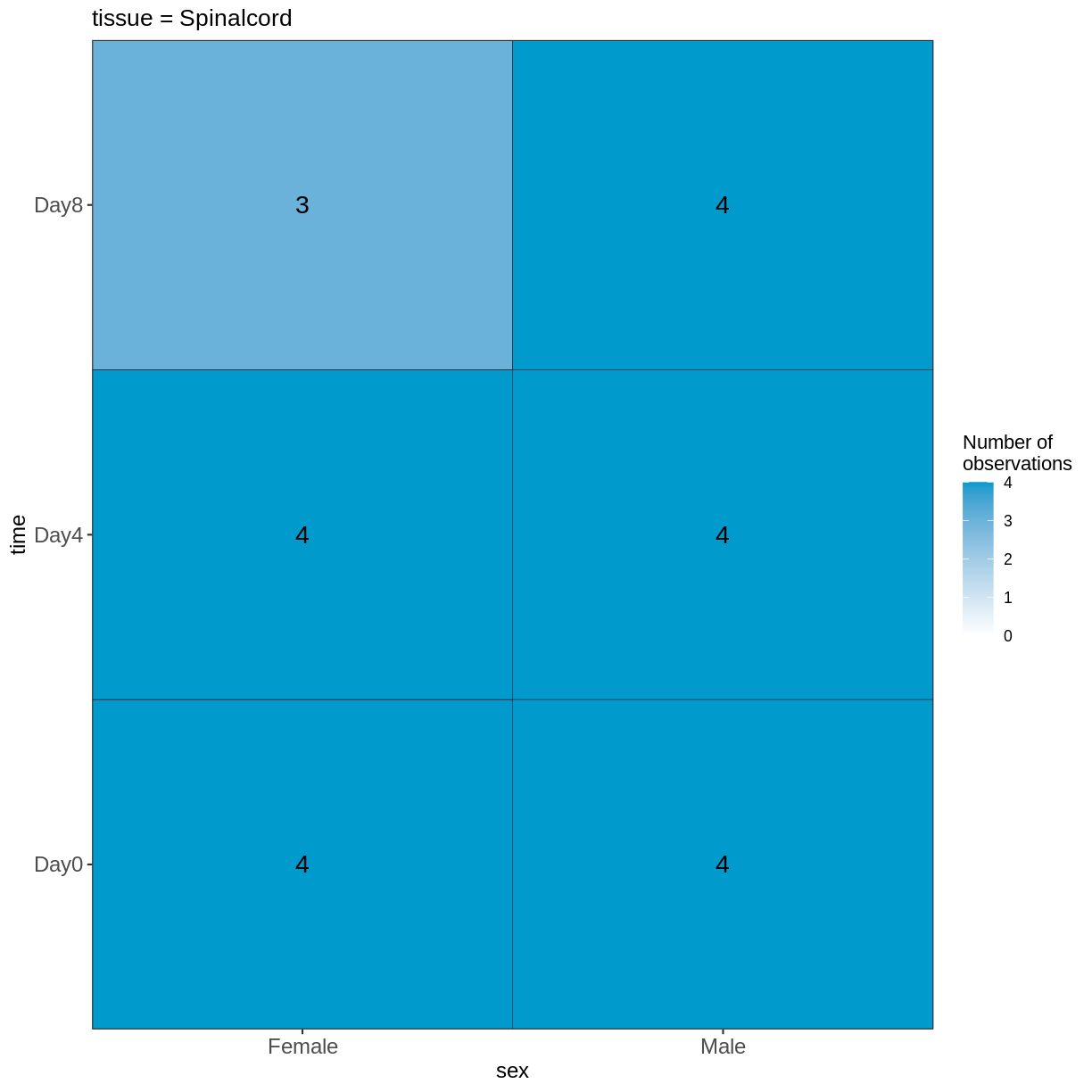
Compare males and females, non-infected spinal cord
meta_noninf_spc <- meta %>% filter(time == "Day0" &
tissue == "Spinalcord")
meta_noninf_spc
title geo_accession organism age sex infection strain time tissue mouse
GSM2545356 CNS_RNA-seq_574 GSM2545356 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 2
GSM2545357 CNS_RNA-seq_575 GSM2545357 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 3
GSM2545358 CNS_RNA-seq_583 GSM2545358 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 4
GSM2545361 CNS_RNA-seq_590 GSM2545361 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 7
GSM2545364 CNS_RNA-seq_709 GSM2545364 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 8
GSM2545365 CNS_RNA-seq_710 GSM2545365 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 9
GSM2545366 CNS_RNA-seq_711 GSM2545366 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 10
GSM2545367 CNS_RNA-seq_713 GSM2545367 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 11
vd <- VisualizeDesign(sampleData = meta_noninf_spc,
designFormula = ~ sex)
vd$designmatrix
(Intercept) sexMale
GSM2545356 1 1
GSM2545357 1 1
GSM2545358 1 0
GSM2545361 1 1
GSM2545364 1 0
GSM2545365 1 0
GSM2545366 1 0
GSM2545367 1 1
vd$plotlist
[[1]]
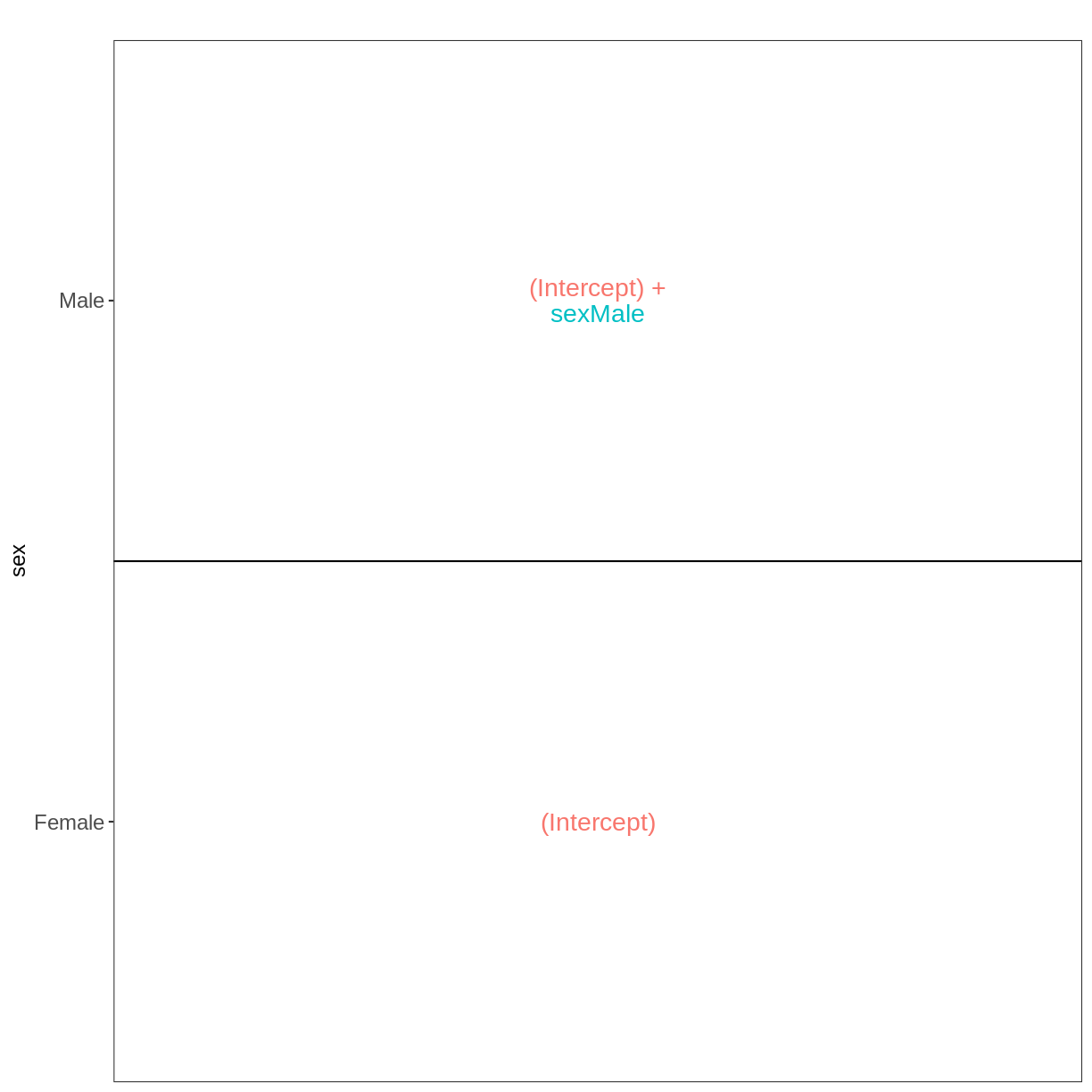
Challenge: Can you do it?
Set up the design formula to compare the three time points (Day0, Day4, Day8) in the male spinal cord samples, and visualize it using
ExploreModelMatrix.Solution
meta_male_spc <- meta %>% filter(sex == "Male" & tissue == "Spinalcord") meta_male_spctitle geo_accession organism age sex infection strain time tissue mouse GSM2545355 CNS_RNA-seq_571 GSM2545355 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 1 GSM2545356 CNS_RNA-seq_574 GSM2545356 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 2 GSM2545357 CNS_RNA-seq_575 GSM2545357 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 3 GSM2545360 CNS_RNA-seq_589 GSM2545360 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 6 GSM2545361 CNS_RNA-seq_590 GSM2545361 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 7 GSM2545367 CNS_RNA-seq_713 GSM2545367 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 11 GSM2545368 CNS_RNA-seq_728 GSM2545368 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 12 GSM2545369 CNS_RNA-seq_729 GSM2545369 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 13 GSM2545372 CNS_RNA-seq_733 GSM2545372 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 17 GSM2545373 CNS_RNA-seq_735 GSM2545373 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day4 Spinalcord 18 GSM2545378 CNS_RNA-seq_742 GSM2545378 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 23 GSM2545379 CNS_RNA-seq_743 GSM2545379 Mus musculus 8 weeks Male InfluenzaA C57BL/6 Day8 Spinalcord 24vd <- VisualizeDesign(sampleData = meta_male_spc, designFormula = ~ time) vd$designmatrix(Intercept) timeDay4 timeDay8 GSM2545355 1 1 0 GSM2545356 1 0 0 GSM2545357 1 0 0 GSM2545360 1 0 1 GSM2545361 1 0 0 GSM2545367 1 0 0 GSM2545368 1 1 0 GSM2545369 1 1 0 GSM2545372 1 0 1 GSM2545373 1 1 0 GSM2545378 1 0 1 GSM2545379 1 0 1vd$plotlist[[1]]
Factorial design without interactions
meta_noninf <- meta %>% filter(time == "Day0")
meta_noninf
title geo_accession organism age sex infection strain time tissue mouse
GSM2545337 CNS_RNA-seq_11C GSM2545337 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 9
GSM2545338 CNS_RNA-seq_12C GSM2545338 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 10
GSM2545343 CNS_RNA-seq_20C GSM2545343 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 11
GSM2545348 CNS_RNA-seq_27C GSM2545348 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 8
GSM2545349 CNS_RNA-seq_28C GSM2545349 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 7
GSM2545353 CNS_RNA-seq_3C GSM2545353 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 4
GSM2545354 CNS_RNA-seq_4C GSM2545354 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 2
GSM2545356 CNS_RNA-seq_574 GSM2545356 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 2
GSM2545357 CNS_RNA-seq_575 GSM2545357 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 3
GSM2545358 CNS_RNA-seq_583 GSM2545358 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 4
GSM2545361 CNS_RNA-seq_590 GSM2545361 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 7
GSM2545364 CNS_RNA-seq_709 GSM2545364 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 8
GSM2545365 CNS_RNA-seq_710 GSM2545365 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 9
GSM2545366 CNS_RNA-seq_711 GSM2545366 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 10
GSM2545367 CNS_RNA-seq_713 GSM2545367 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 11
vd <- VisualizeDesign(sampleData = meta_noninf,
designFormula = ~ sex + tissue)
vd$designmatrix
(Intercept) sexMale tissueSpinalcord
GSM2545337 1 0 0
GSM2545338 1 0 0
GSM2545343 1 1 0
GSM2545348 1 0 0
GSM2545349 1 1 0
GSM2545353 1 0 0
GSM2545354 1 1 0
GSM2545356 1 1 1
GSM2545357 1 1 1
GSM2545358 1 0 1
GSM2545361 1 1 1
GSM2545364 1 0 1
GSM2545365 1 0 1
GSM2545366 1 0 1
GSM2545367 1 1 1
vd$plotlist
[[1]]
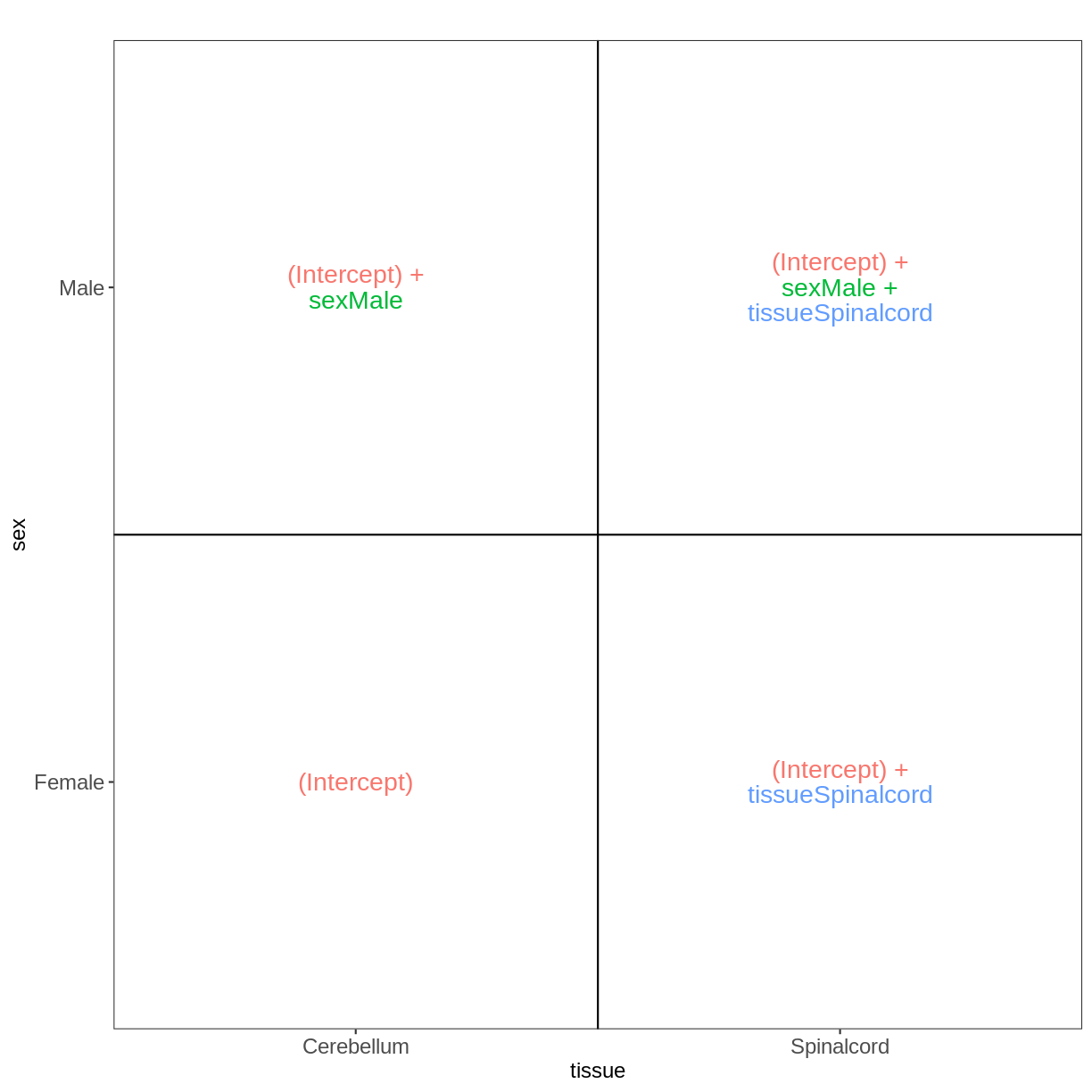
Factorial design with interactions
meta_noninf <- meta %>% filter(time == "Day0")
meta_noninf
title geo_accession organism age sex infection strain time tissue mouse
GSM2545337 CNS_RNA-seq_11C GSM2545337 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 9
GSM2545338 CNS_RNA-seq_12C GSM2545338 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 10
GSM2545343 CNS_RNA-seq_20C GSM2545343 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 11
GSM2545348 CNS_RNA-seq_27C GSM2545348 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 8
GSM2545349 CNS_RNA-seq_28C GSM2545349 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 7
GSM2545353 CNS_RNA-seq_3C GSM2545353 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 4
GSM2545354 CNS_RNA-seq_4C GSM2545354 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Cerebellum 2
GSM2545356 CNS_RNA-seq_574 GSM2545356 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 2
GSM2545357 CNS_RNA-seq_575 GSM2545357 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 3
GSM2545358 CNS_RNA-seq_583 GSM2545358 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 4
GSM2545361 CNS_RNA-seq_590 GSM2545361 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 7
GSM2545364 CNS_RNA-seq_709 GSM2545364 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 8
GSM2545365 CNS_RNA-seq_710 GSM2545365 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 9
GSM2545366 CNS_RNA-seq_711 GSM2545366 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 10
GSM2545367 CNS_RNA-seq_713 GSM2545367 Mus musculus 8 weeks Male NonInfected C57BL/6 Day0 Spinalcord 11
vd <- VisualizeDesign(sampleData = meta_noninf,
designFormula = ~ sex * tissue)
vd$designmatrix
(Intercept) sexMale tissueSpinalcord sexMale:tissueSpinalcord
GSM2545337 1 0 0 0
GSM2545338 1 0 0 0
GSM2545343 1 1 0 0
GSM2545348 1 0 0 0
GSM2545349 1 1 0 0
GSM2545353 1 0 0 0
GSM2545354 1 1 0 0
GSM2545356 1 1 1 1
GSM2545357 1 1 1 1
GSM2545358 1 0 1 0
GSM2545361 1 1 1 1
GSM2545364 1 0 1 0
GSM2545365 1 0 1 0
GSM2545366 1 0 1 0
GSM2545367 1 1 1 1
vd$plotlist
[[1]]
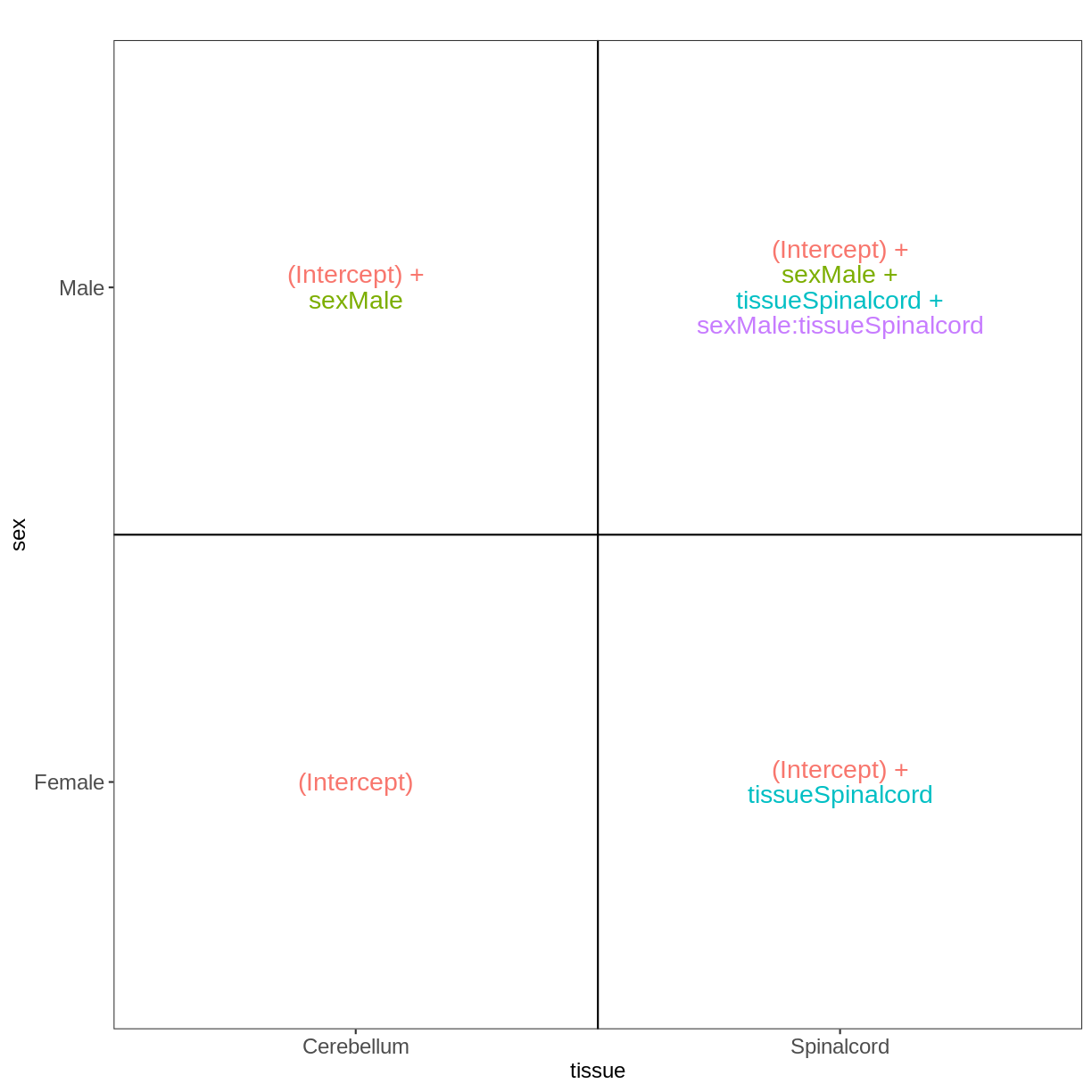
Paired design
meta_fem_day0 <- meta %>% filter(sex == "Female" &
time == "Day0")
meta_fem_day0
title geo_accession organism age sex infection strain time tissue mouse
GSM2545337 CNS_RNA-seq_11C GSM2545337 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 9
GSM2545338 CNS_RNA-seq_12C GSM2545338 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 10
GSM2545348 CNS_RNA-seq_27C GSM2545348 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 8
GSM2545353 CNS_RNA-seq_3C GSM2545353 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 4
GSM2545358 CNS_RNA-seq_583 GSM2545358 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 4
GSM2545364 CNS_RNA-seq_709 GSM2545364 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 8
GSM2545365 CNS_RNA-seq_710 GSM2545365 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 9
GSM2545366 CNS_RNA-seq_711 GSM2545366 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 10
vd <- VisualizeDesign(sampleData = meta_fem_day0,
designFormula = ~ mouse + tissue)
vd$designmatrix
(Intercept) mouse tissueSpinalcord
GSM2545337 1 9 0
GSM2545338 1 10 0
GSM2545348 1 8 0
GSM2545353 1 4 0
GSM2545358 1 4 1
GSM2545364 1 8 1
GSM2545365 1 9 1
GSM2545366 1 10 1
vd$plotlist
[[1]]
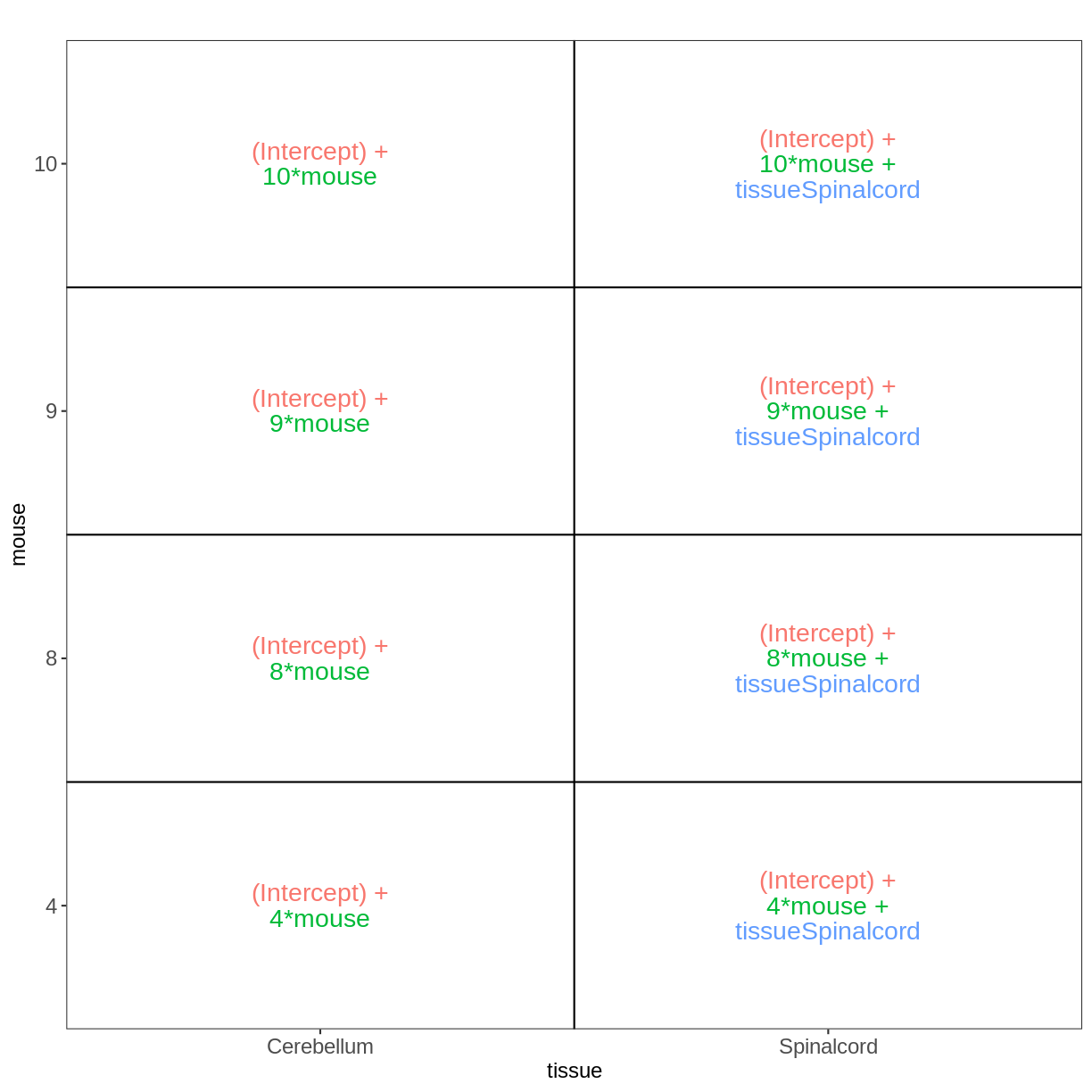
Within- and between-subject comparisons
meta_fem_day04 <- meta %>%
filter(sex == "Female" &
time %in% c("Day0", "Day4")) %>%
droplevels()
meta_fem_day04
title geo_accession organism age sex infection strain time tissue mouse
GSM2545337 CNS_RNA-seq_11C GSM2545337 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 9
GSM2545338 CNS_RNA-seq_12C GSM2545338 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 10
GSM2545339 CNS_RNA-seq_13C GSM2545339 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 15
GSM2545344 CNS_RNA-seq_21C GSM2545344 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 22
GSM2545348 CNS_RNA-seq_27C GSM2545348 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 8
GSM2545352 CNS_RNA-seq_30C GSM2545352 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 21
GSM2545353 CNS_RNA-seq_3C GSM2545353 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Cerebellum 4
GSM2545358 CNS_RNA-seq_583 GSM2545358 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 4
GSM2545362 CNS_RNA-seq_5C GSM2545362 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Cerebellum 20
GSM2545364 CNS_RNA-seq_709 GSM2545364 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 8
GSM2545365 CNS_RNA-seq_710 GSM2545365 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 9
GSM2545366 CNS_RNA-seq_711 GSM2545366 Mus musculus 8 weeks Female NonInfected C57BL/6 Day0 Spinalcord 10
GSM2545371 CNS_RNA-seq_731 GSM2545371 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 15
GSM2545375 CNS_RNA-seq_738 GSM2545375 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 20
GSM2545376 CNS_RNA-seq_740 GSM2545376 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 21
GSM2545377 CNS_RNA-seq_741 GSM2545377 Mus musculus 8 weeks Female InfluenzaA C57BL/6 Day4 Spinalcord 22
design <- model.matrix(~ mouse, data = meta_fem_day04)
design <- cbind(design,
Spc.Day0 = meta_fem_day04$tissue == "Spinalcord" &
meta_fem_day04$time == "Day0",
Spc.Day4 = meta_fem_day04$tissue == "Spinalcord" &
meta_fem_day04$time == "Day4")
rownames(design) <- rownames(meta_fem_day04)
design
(Intercept) mouse Spc.Day0 Spc.Day4
GSM2545337 1 9 0 0
GSM2545338 1 10 0 0
GSM2545339 1 15 0 0
GSM2545344 1 22 0 0
GSM2545348 1 8 0 0
GSM2545352 1 21 0 0
GSM2545353 1 4 0 0
GSM2545358 1 4 1 0
GSM2545362 1 20 0 0
GSM2545364 1 8 1 0
GSM2545365 1 9 1 0
GSM2545366 1 10 1 0
GSM2545371 1 15 0 1
GSM2545375 1 20 0 1
GSM2545376 1 21 0 1
GSM2545377 1 22 0 1
vd <- VisualizeDesign(sampleData = meta_fem_day04 %>%
select(time, tissue, mouse),
designFormula = NULL,
designMatrix = design, flipCoordFitted = FALSE)
vd$designmatrix
(Intercept) mouse Spc.Day0 Spc.Day4
GSM2545337 1 9 0 0
GSM2545338 1 10 0 0
GSM2545339 1 15 0 0
GSM2545344 1 22 0 0
GSM2545348 1 8 0 0
GSM2545352 1 21 0 0
GSM2545353 1 4 0 0
GSM2545358 1 4 1 0
GSM2545362 1 20 0 0
GSM2545364 1 8 1 0
GSM2545365 1 9 1 0
GSM2545366 1 10 1 0
GSM2545371 1 15 0 1
GSM2545375 1 20 0 1
GSM2545376 1 21 0 1
GSM2545377 1 22 0 1
vd$plotlist
$`time = Day0`
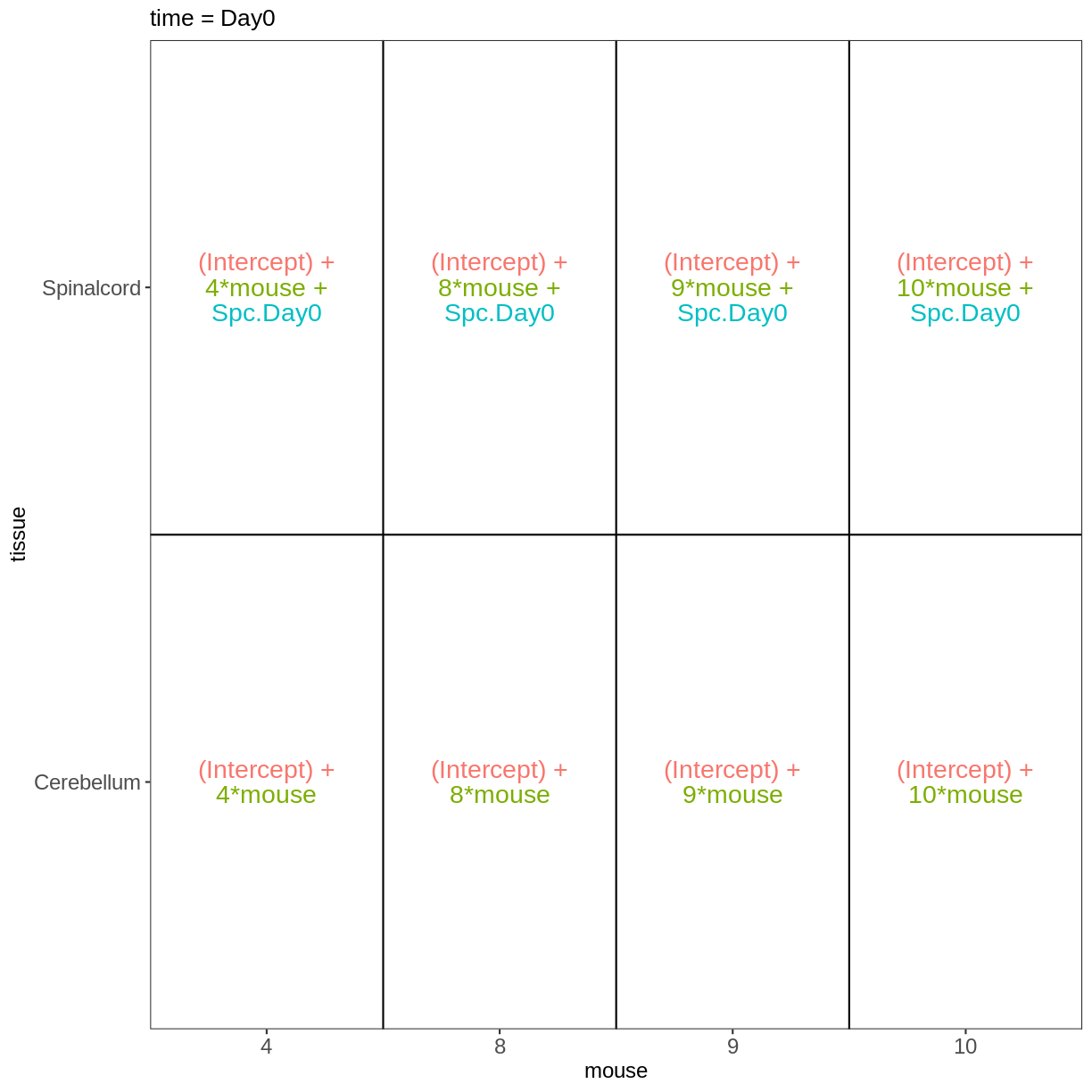
$`time = Day4`
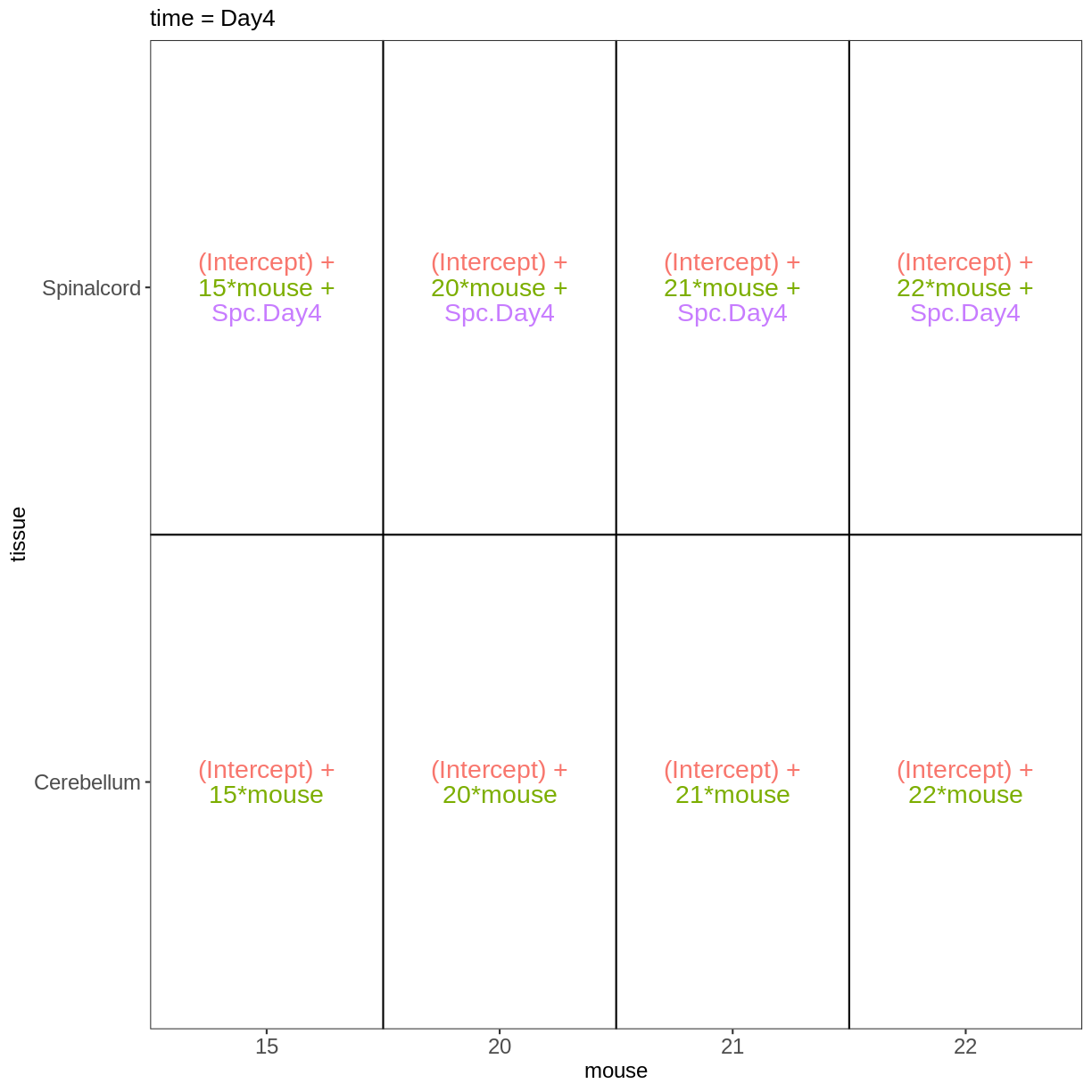
Key Points
Key point 1
Importing and annotating quantified data into R
Overview
Teaching: XX min
Exercises: XX minQuestions
How do we get our data into R?
Objectives
Learn how to import the quantifications into a SummarizedExperiment object.
Learn how to add additional gene annotations to the object.
Contribute!
This episode is intended to show how we can assemble a SummarizedExperiment starting from individual count, rowdata and coldata files. Moreover, we will practice adding annotations for the genes, and discuss related concepts and things to keep in mind (annotation sources, versions, ‘helper’ packages like tximeta).
Read the data
Counts
counts <- read.csv("data/GSE96870_counts_cerebellum.csv",
row.names = 1)
Sample annotations
coldata <- read.csv("data/GSE96870_coldata_cerebellum.csv",
row.names = 1)
Gene annotations
Need to be careful - the descriptions contain both commas and ‘ (e.g., 5’)
rowranges <- read.delim("data/GSE96870_rowranges.tsv", sep = "\t",
colClasses = c(ENTREZID = "character"),
header = TRUE, quote = "", row.names = 5)
Mention other ways of getting annotations, and practice querying org package. Important to use the right annotation source/version.
suppressPackageStartupMessages({
library(org.Mm.eg.db)
})
mapIds(org.Mm.eg.db, keys = "497097", column = "SYMBOL", keytype = "ENTREZID")
'select()' returned 1:1 mapping between keys and columns
497097
"Xkr4"
Check feature types
table(rowranges$gbkey)
C_region D_segment exon J_segment misc_RNA
20 23 4008 94 1988
mRNA ncRNA precursor_RNA rRNA tRNA
21198 12285 1187 35 413
V_segment
535
Assemble SummarizedExperiment
stopifnot(rownames(rowranges) == rownames(counts),
rownames(coldata) == colnames(counts))
se <- SummarizedExperiment(
assays = list(counts = as.matrix(counts)),
rowRanges = as(rowranges, "GRanges"),
colData = coldata
)
Save SummarizedExperiment
saveRDS(se, "data/GSE96870_se.rds")
Session info
sessionInfo()
R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.2 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] org.Mm.eg.db_3.13.0 AnnotationDbi_1.54.1
[3] knitr_1.33 SummarizedExperiment_1.22.0
[5] Biobase_2.52.0 MatrixGenerics_1.4.0
[7] matrixStats_0.60.0 GenomicRanges_1.44.0
[9] GenomeInfoDb_1.28.1 IRanges_2.26.0
[11] S4Vectors_0.30.0 BiocGenerics_0.38.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 compiler_4.1.0 XVector_0.32.0
[4] bitops_1.0-7 tools_4.1.0 zlibbioc_1.38.0
[7] bit_4.0.4 evaluate_0.14 RSQLite_2.2.7
[10] memoise_2.0.0 lattice_0.20-44 pkgconfig_2.0.3
[13] png_0.1-7 rlang_0.4.11 Matrix_1.3-3
[16] DelayedArray_0.18.0 DBI_1.1.1 xfun_0.24
[19] fastmap_1.1.0 GenomeInfoDbData_1.2.6 stringr_1.4.0
[22] httr_1.4.2 Biostrings_2.60.1 vctrs_0.3.8
[25] bit64_4.0.5 grid_4.1.0 R6_2.5.0
[28] blob_1.2.2 magrittr_2.0.1 KEGGREST_1.32.0
[31] stringi_1.7.3 RCurl_1.98-1.3 cachem_1.0.5
[34] crayon_1.4.1
Key Points
Key point 1
Exploratory analysis and quality control
Overview
Teaching: XX min
Exercises: XX minQuestions
Learn how to explore the gene expression matrix and perform common quality control steps.
Learn how to set up an interactive application for exploratory analysis.
Contribute!
This episode is intended to introduce various types of exploratory analysis and QC steps taken before a formal statistical analysis is done.
suppressPackageStartupMessages({
library(SummarizedExperiment)
library(DESeq2)
library(vsn)
library(ggplot2)
library(ComplexHeatmap)
library(RColorBrewer)
library(hexbin)
})
se <- readRDS("data/GSE96870_se.rds")
Exploratory analysis is crucial for quality control and to get to know our data. It can help us detect quality problems, sample swaps and contamination, as well as give us a sense of the most salient patterns present in the data. In this episode, we will learn about two common ways of performing exploratory analysis for RNA-seq data; namely clustering and principal component analysis (PCA). These tools are in no way limited to (or developed for) analysis of RNA-seq data. However, there are certain characteristics of count assays that need to be taken into account when they are applied to this type of data.
se <- se[rowSums(assay(se, "counts")) > 5, ]
dds <- DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"],
design = ~ sex + time)
Warning in DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"], design = ~sex
+ : some variables in design formula are characters, converting to factors
Library size differences
ggplot(data.frame(sample = colnames(dds),
libSize = colSums(assay(dds, "counts"))),
aes(x = sample, y = libSize)) +
geom_bar(stat = "identity") + theme_bw() +
labs(x = "Sample", y = "Total count") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))
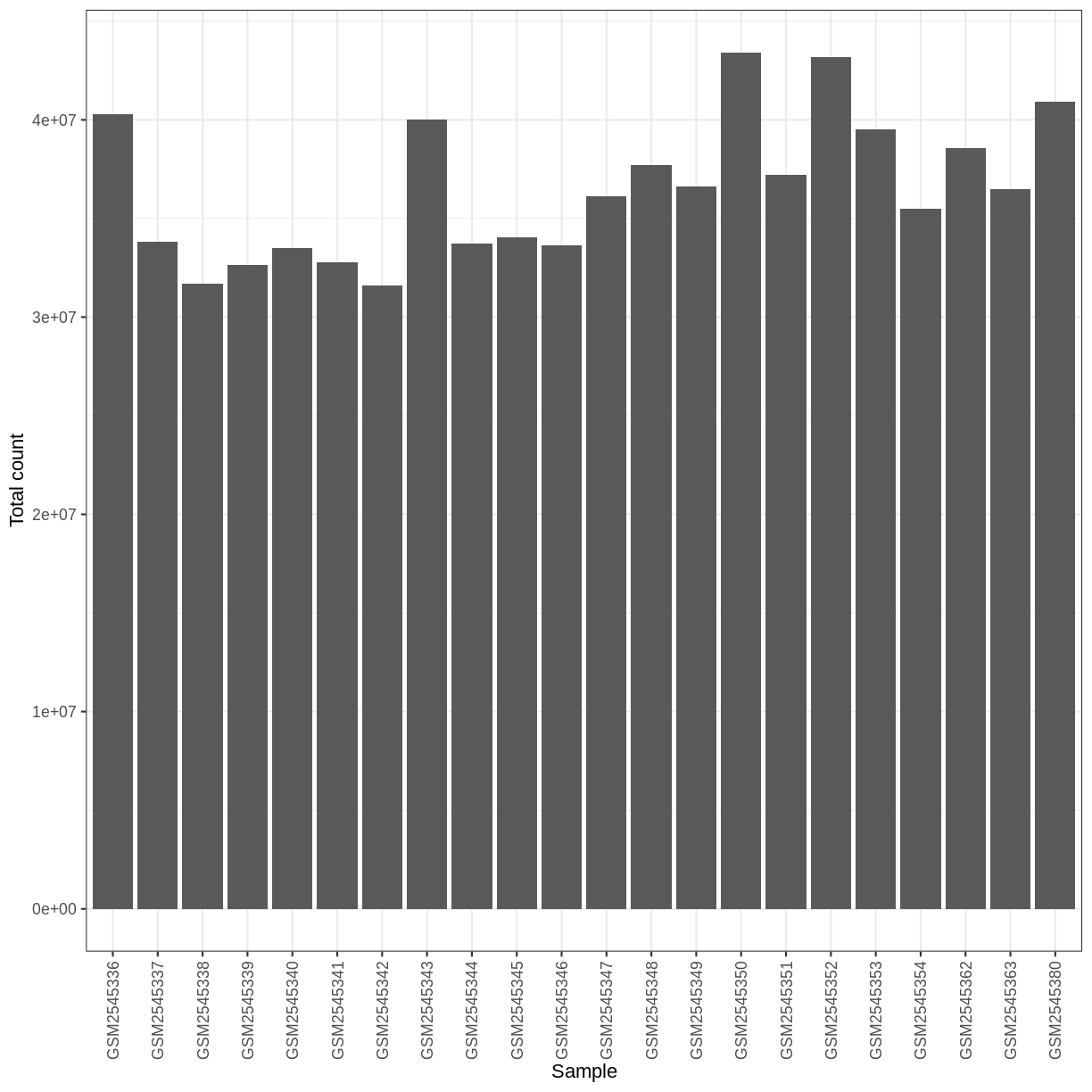
Differences in the total number of reads assigned to genes between samples typically occur for technical reasons. In practice, it means that we can not simply compare the raw read count directly between samples and conclude that a sample with a higher read count also expresses the gene more strongly - the higher count may be caused by an overall higher number of reads in that sample. In the rest of this section, we will use the term library size to refer to the total number of reads assigned to genes for a sample. We need to adjust for the differences in library size between samples, to avoid drawing incorrect conclusions. The way this is typically done for RNA-seq data can be described as a two-step procedure. First, we estimate size factors - sample-specific correction factors such that if the raw counts were to be divided by these factors, the resulting values would be more comparable across samples. Next, these size factors are incorporated into the statistical analysis of the data. It is important to pay close attention to how this is done in practice for a given analysis method. Sometimes the division of the counts by the size factors needs to be done explicitly by the analyst. Other times (as we will see for the differential expression analysis) it is important that they are provided separately to the analysis tool, which will then use them appropriately in the statistical model.
With DESeq2, size factors are calculated using the estimateSizeFactors() function.
The size factors estimated by this function combines an adjustment for differences in library sizes with an adjustment for differences in the RNA composition of the samples.
The latter is important due to the compositional nature of RNA-seq data.
There is a fixed number of reads to distribute between the genes, and if a single (or a few) very highly expressed gene consume a large part of the reads, all other genes will consequently receive very low counts.
dds <- estimateSizeFactors(dds)
ggplot(data.frame(libSize = colSums(assay(dds, "counts")),
sizeFactor = sizeFactors(dds)),
aes(x = libSize, y = sizeFactor)) +
geom_point(size = 5) + theme_bw() +
labs(x = "Library size", y = "Size factor")
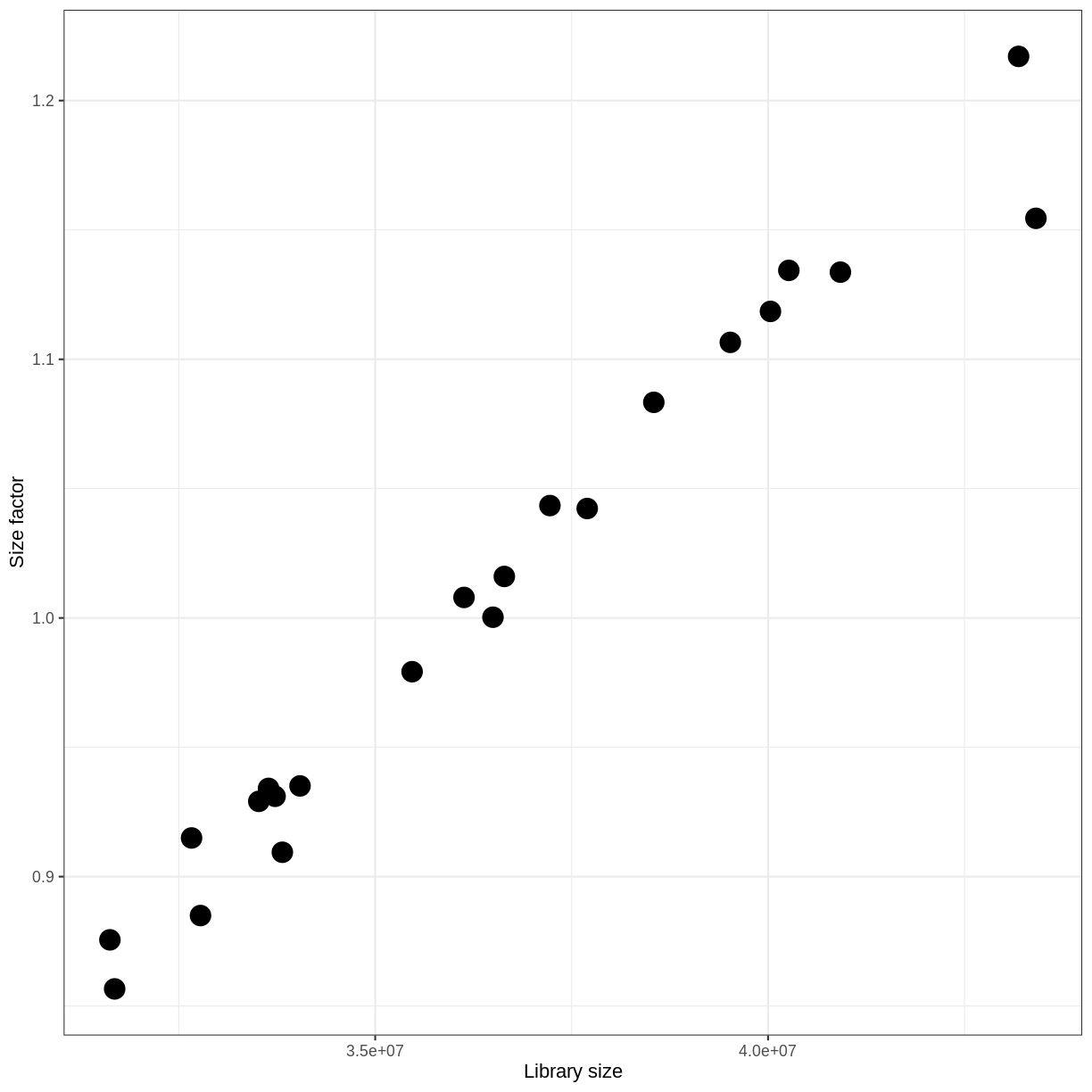
Transform data
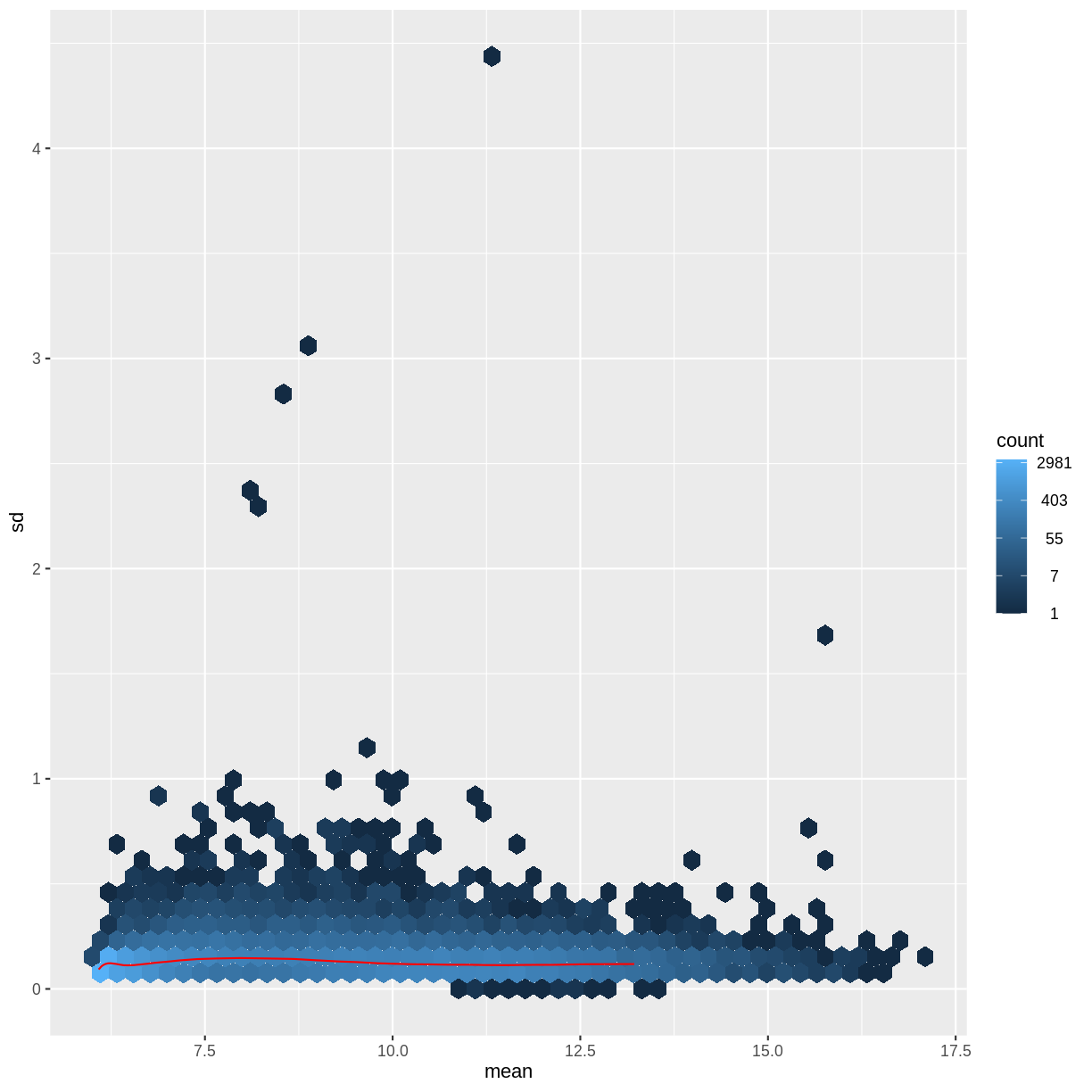
There is a rich literature on methods for exploratory analysis. Most of these work best in situations where the variance of the input data (here, each gene) is relatively independent of the average value. For read count data such as RNA-seq, this is not the case. In fact, the variance increases with the average read count.
meanSdPlot(assay(dds), ranks = FALSE)
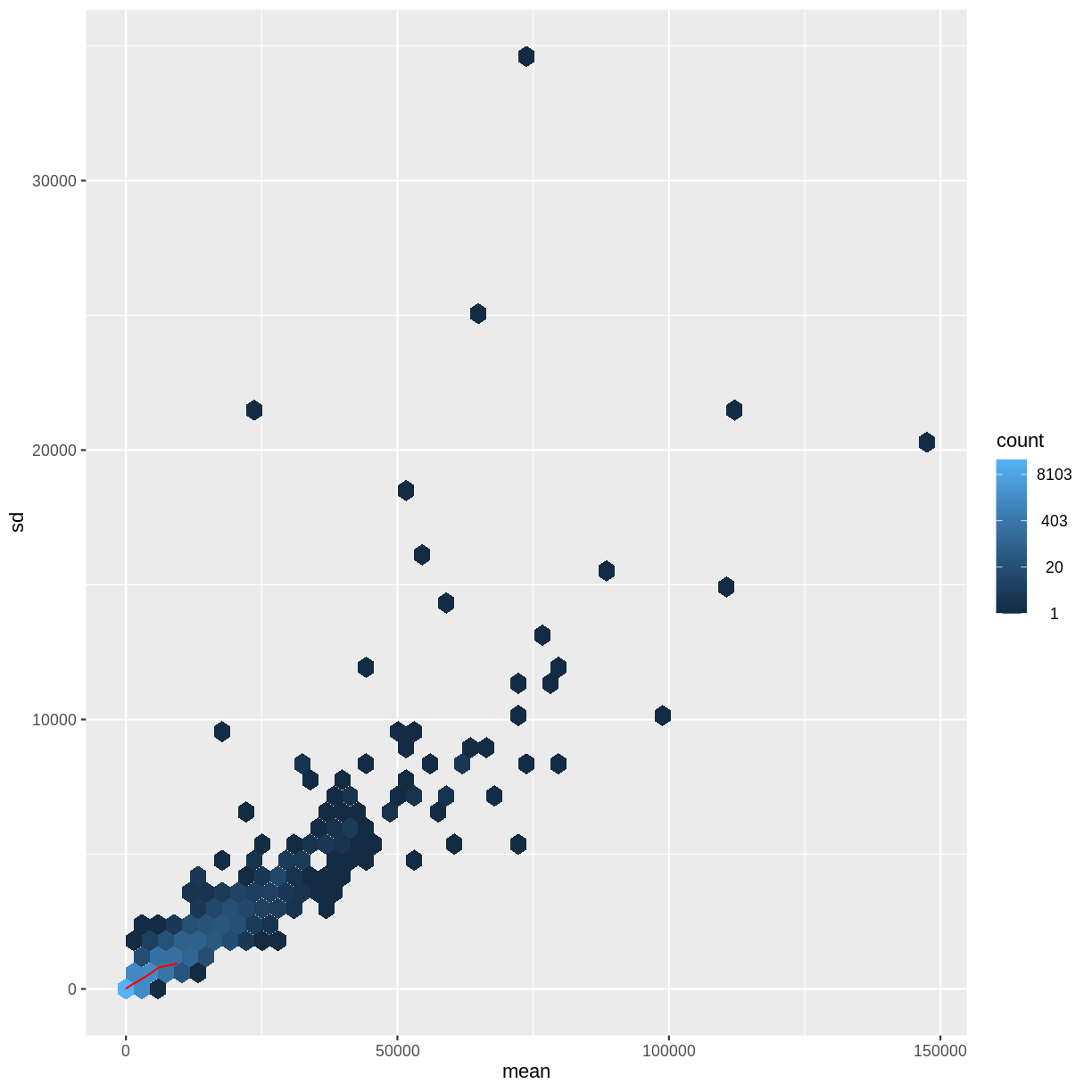
There are two ways around this: either we develop methods specifically adapted to count data, or we adapt (transform) the count data so that the existing methods are applicable. Both ways have been explored; however, at the moment the second approach is arguably more widely applied in practice.
vsd <- DESeq2::vst(dds, blind = TRUE)
meanSdPlot(assay(vsd), ranks = FALSE)
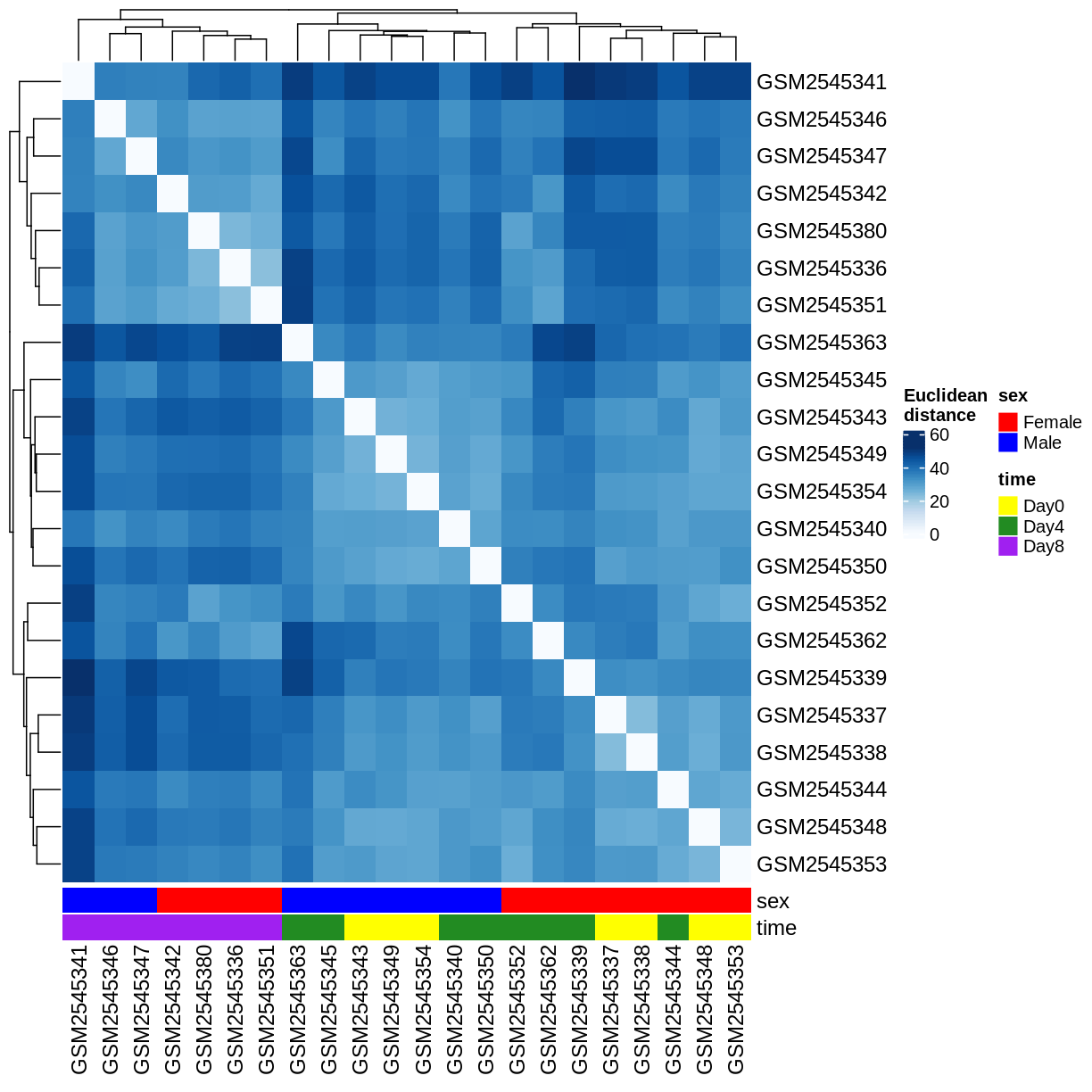
Heatmaps and clustering
dst <- dist(t(assay(vsd)))
colors <- colorRampPalette(brewer.pal(9, "Blues"))(255)
ComplexHeatmap::Heatmap(
as.matrix(dst),
col = colors,
name = "Euclidean\ndistance",
cluster_rows = hclust(dst),
cluster_columns = hclust(dst),
bottom_annotation = columnAnnotation(
sex = vsd$sex,
time = vsd$time,
col = list(sex = c(Female = "red", Male = "blue"),
time = c(Day0 = "yellow", Day4 = "forestgreen", Day8 = "purple")))
)
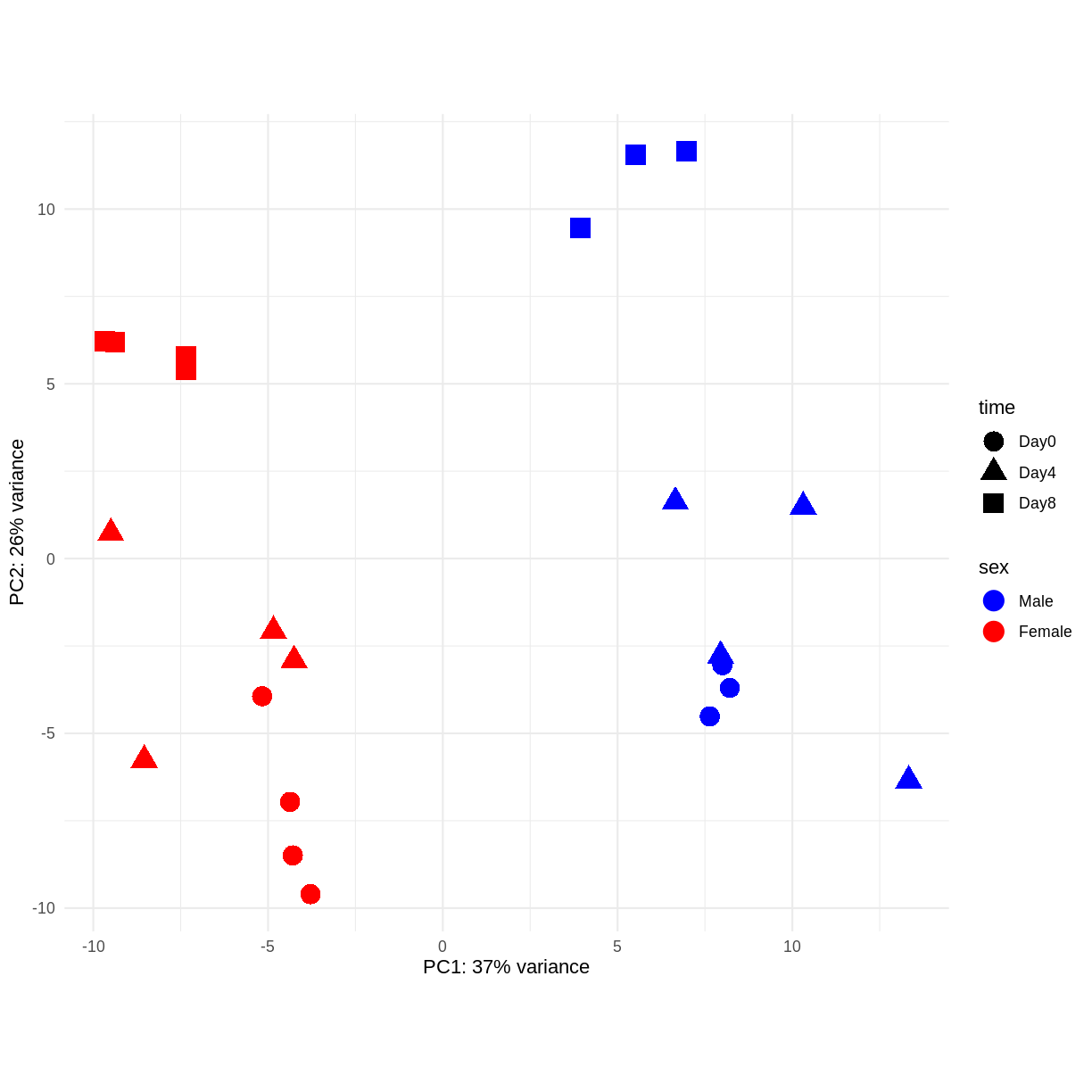
PCA
Principal component analysis is a dimensionality reduction method, which projects the samples into a lower-dimensional space. This lower-dimensional representation can be used for visualization, or as the input for other analysis methods. The principal components are defined in such a way that they are orthogonal, and that the projection of the samples into the space they span contains as much variance as possible. It is an unsupervised method in the sense that no external information about the samples (e.g., the treatment condition) is taken into account. In the plot below we represent the samples in a two-dimensional principal component space. For each of the two dimensions, we indicate the fraction of the total variance that is represented by that component. By definition, the first principal component will always represent more of the variance than the subsequent ones. The fraction of explained variance is a measure of how much of the ‘signal’ in the data that is retained when we project the samples from the original, high-dimensional space to the low-dimensional space for visualization.
pcaData <- DESeq2::plotPCA(vsd, intgroup = c("sex", "time"),
returnData = TRUE)
percentVar <- round(100 * attr(pcaData, "percentVar"))
ggplot(pcaData, aes(x = PC1, y = PC2)) +
geom_point(aes(color = sex, shape = time), size = 5) +
theme_minimal() +
xlab(paste0("PC1: ", percentVar[1], "% variance")) +
ylab(paste0("PC2: ", percentVar[2], "% variance")) +
coord_fixed() +
scale_color_manual(values = c(Male = "blue", Female = "red"))
Session info
sessionInfo()
R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.2 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] hexbin_1.28.2 RColorBrewer_1.1-2
[3] ComplexHeatmap_2.8.0 ggplot2_3.3.5
[5] vsn_3.60.0 DESeq2_1.32.0
[7] knitr_1.33 SummarizedExperiment_1.22.0
[9] Biobase_2.52.0 MatrixGenerics_1.4.0
[11] matrixStats_0.60.0 GenomicRanges_1.44.0
[13] GenomeInfoDb_1.28.1 IRanges_2.26.0
[15] S4Vectors_0.30.0 BiocGenerics_0.38.0
loaded via a namespace (and not attached):
[1] httr_1.4.2 foreach_1.5.1 bit64_4.0.5
[4] splines_4.1.0 highr_0.9 BiocManager_1.30.16
[7] affy_1.70.0 blob_1.2.2 GenomeInfoDbData_1.2.6
[10] pillar_1.6.2 RSQLite_2.2.7 lattice_0.20-44
[13] glue_1.4.2 limma_3.48.1 digest_0.6.27
[16] XVector_0.32.0 colorspace_2.0-2 preprocessCore_1.54.0
[19] Matrix_1.3-3 XML_3.99-0.6 pkgconfig_2.0.3
[22] GetoptLong_1.0.5 genefilter_1.74.0 zlibbioc_1.38.0
[25] purrr_0.3.4 xtable_1.8-4 scales_1.1.1
[28] affyio_1.62.0 BiocParallel_1.26.1 tibble_3.1.3
[31] annotate_1.70.0 KEGGREST_1.32.0 farver_2.1.0
[34] generics_0.1.0 ellipsis_0.3.2 cachem_1.0.5
[37] withr_2.4.2 survival_3.2-11 magrittr_2.0.1
[40] crayon_1.4.1 memoise_2.0.0 evaluate_0.14
[43] fansi_0.5.0 doParallel_1.0.16 Cairo_1.5-12.2
[46] tools_4.1.0 GlobalOptions_0.1.2 lifecycle_1.0.0
[49] stringr_1.4.0 munsell_0.5.0 locfit_1.5-9.4
[52] cluster_2.1.2 DelayedArray_0.18.0 AnnotationDbi_1.54.1
[55] Biostrings_2.60.1 compiler_4.1.0 rlang_0.4.11
[58] RCurl_1.98-1.3 iterators_1.0.13 circlize_0.4.13
[61] rjson_0.2.20 labeling_0.4.2 bitops_1.0-7
[64] codetools_0.2-18 gtable_0.3.0 DBI_1.1.1
[67] R6_2.5.0 dplyr_1.0.7 fastmap_1.1.0
[70] bit_4.0.4 utf8_1.2.2 clue_0.3-59
[73] shape_1.4.6 stringi_1.7.3 Rcpp_1.0.7
[76] vctrs_0.3.8 geneplotter_1.70.0 png_0.1-7
[79] tidyselect_1.1.1 xfun_0.24
Key Points
Key point 1
Differential expression analysis
Overview
Teaching: XX min
Exercises: XX minQuestions
How do we find differentially expressed genes?
Objectives
Explain the steps involved in a differential expression analysis.
Explain how to perform these steps in R, using DESeq2.
Contribute!
This episode is intended to introduce the concepts required to perform differential expression analysis with RNA-seq data. Explain concepts like size factors, count modeling (Negative Binomial), dispersion, interpretation of the test output, multiple testing correction.
suppressPackageStartupMessages({
library(SummarizedExperiment)
library(DESeq2)
library(ggplot2)
library(ExploreModelMatrix)
library(cowplot)
library(ComplexHeatmap)
})
Load data
se <- readRDS("data/GSE96870_se.rds")
Create DESeqDataSet
dds <- DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"],
design = ~ sex + time)
Warning in DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"], design = ~sex + : some variables in design formula are
characters, converting to factors
Run DESeq()
Contribute!
The concepts may be clearer if the steps of DESeq() are first performed separately, followed by a note that they can be performed in a single step using DESeq().
dds <- DESeq2::DESeq(dds)
estimating size factors
estimating dispersions
gene-wise dispersion estimates
mean-dispersion relationship
final dispersion estimates
fitting model and testing
plotDispEsts(dds)
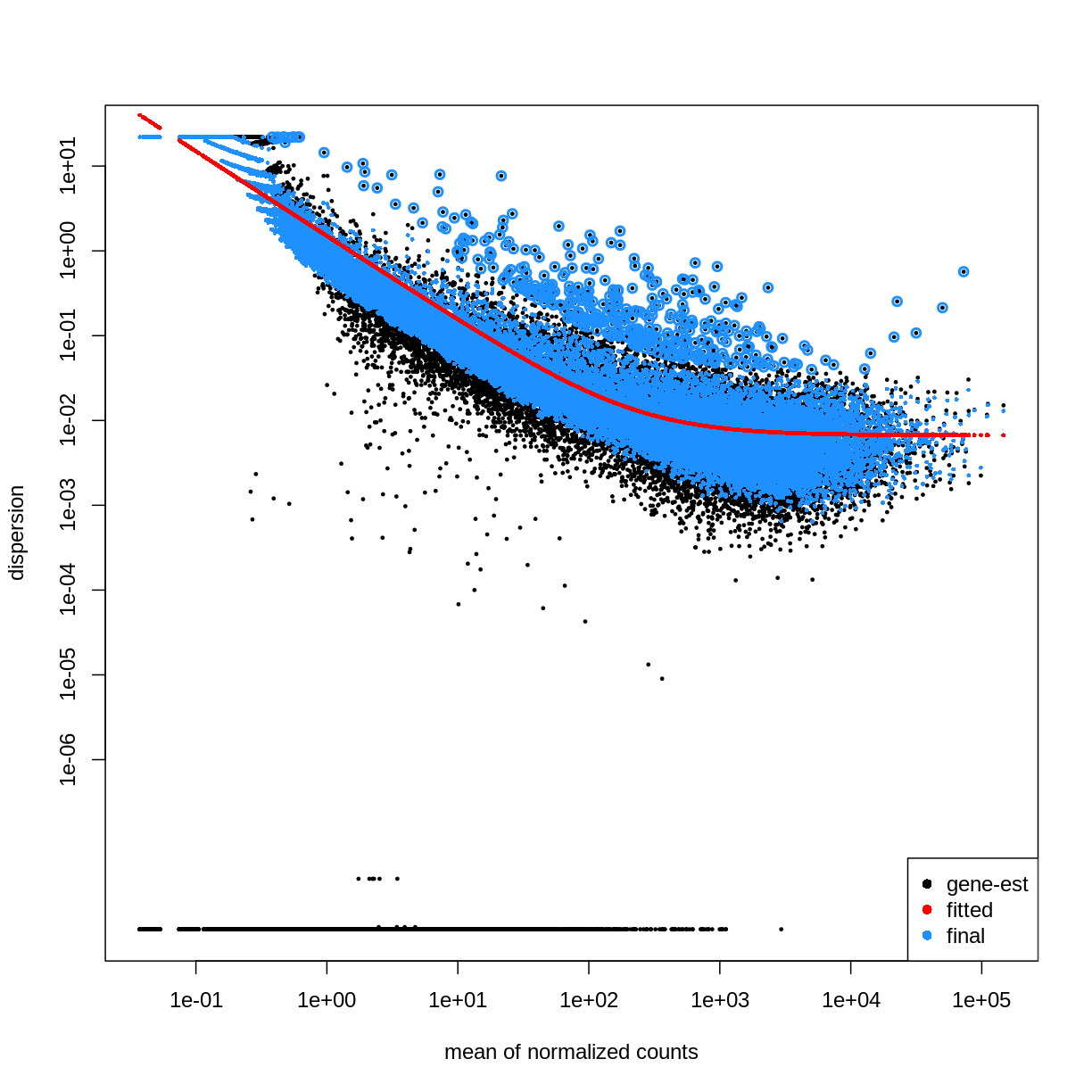
Extract results for specific contrasts
Contribute!
Refer back to the episode about experimental design.
## Day 8 vs Day 0
resTime <- DESeq2::results(dds, contrast = c("time", "Day8", "Day0"))
summary(resTime)
out of 32652 with nonzero total read count
adjusted p-value < 0.1
LFC > 0 (up) : 4472, 14%
LFC < 0 (down) : 4276, 13%
outliers [1] : 10, 0.031%
low counts [2] : 8732, 27%
(mean count < 1)
[1] see 'cooksCutoff' argument of ?results
[2] see 'independentFiltering' argument of ?results
head(resTime[order(resTime$pvalue), ])
log2 fold change (MLE): time Day8 vs Day0
Wald test p-value: time Day8 vs Day0
DataFrame with 6 rows and 6 columns
baseMean log2FoldChange lfcSE stat pvalue padj
<numeric> <numeric> <numeric> <numeric> <numeric> <numeric>
Asl 701.343 1.11733 0.0592541 18.8565 2.59885e-79 6.21386e-75
Apod 18765.146 1.44698 0.0805186 17.9708 3.30147e-72 3.94690e-68
Cyp2d22 2550.480 0.91020 0.0554756 16.4072 1.69794e-60 1.35326e-56
Klk6 546.503 -1.67190 0.1058989 -15.7877 3.78228e-56 2.26086e-52
Fcrls 184.235 -1.94701 0.1279847 -15.2128 2.90708e-52 1.39017e-48
A330076C08Rik 107.250 -1.74995 0.1154279 -15.1606 6.45112e-52 2.57077e-48
DESeq2::plotMA(resTime)
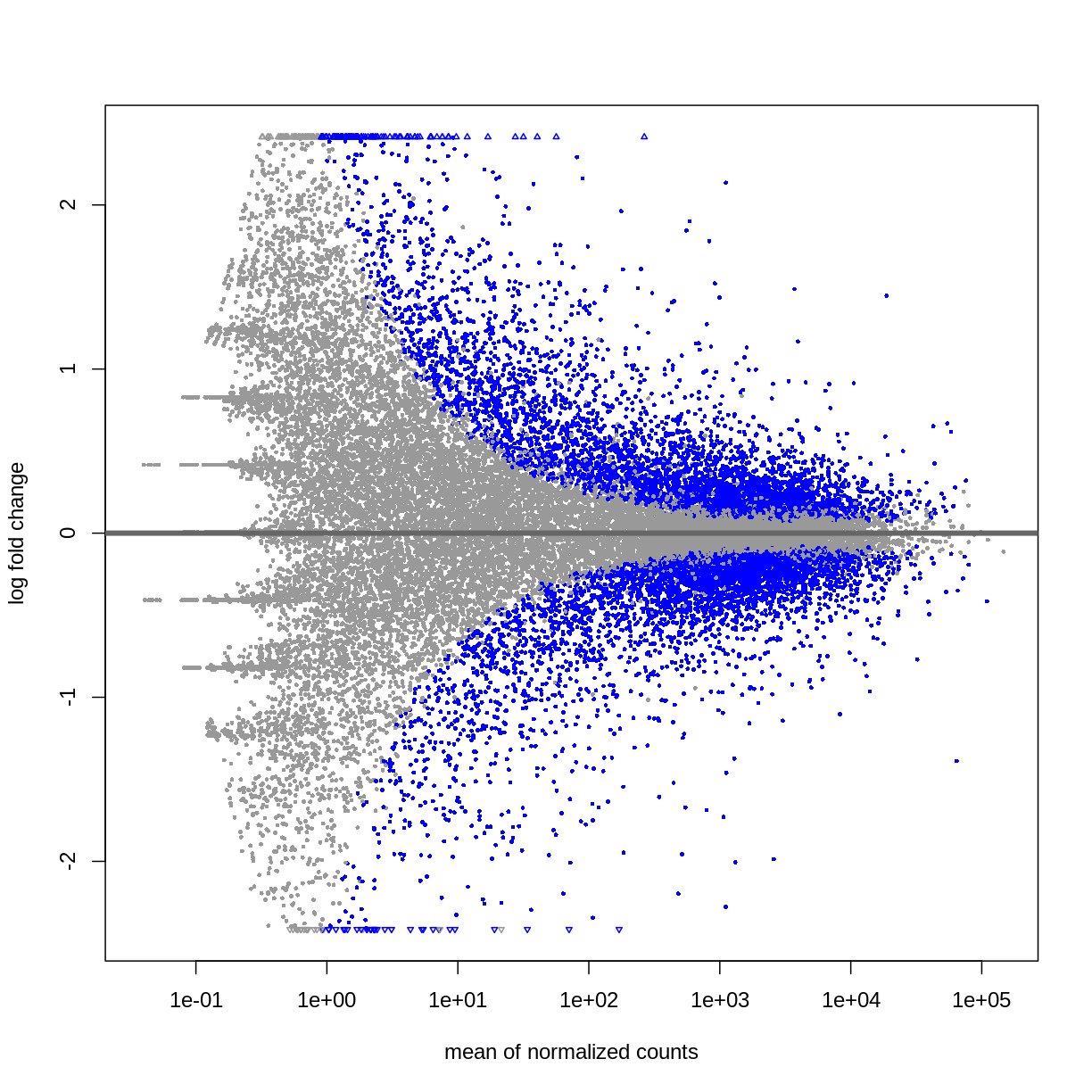
## Male vs Female
resSex <- DESeq2::results(dds, contrast = c("sex", "Male", "Female"))
summary(resSex)
out of 32652 with nonzero total read count
adjusted p-value < 0.1
LFC > 0 (up) : 53, 0.16%
LFC < 0 (down) : 71, 0.22%
outliers [1] : 10, 0.031%
low counts [2] : 13717, 42%
(mean count < 6)
[1] see 'cooksCutoff' argument of ?results
[2] see 'independentFiltering' argument of ?results
head(resSex[order(resSex$pvalue), ])
log2 fold change (MLE): sex Male vs Female
Wald test p-value: sex Male vs Female
DataFrame with 6 rows and 6 columns
baseMean log2FoldChange lfcSE stat pvalue padj
<numeric> <numeric> <numeric> <numeric> <numeric> <numeric>
Xist 22603.0359 -11.60429 0.336282 -34.5076 6.16852e-261 1.16739e-256
Ddx3y 2072.9436 11.87241 0.397493 29.8683 5.08722e-196 4.81378e-192
Eif2s3y 1410.8750 12.62514 0.565216 22.3369 1.62066e-110 1.02237e-106
Kdm5d 692.1672 12.55386 0.593627 21.1477 2.89566e-99 1.37001e-95
Uty 667.4375 12.01728 0.593591 20.2451 3.92780e-91 1.48667e-87
LOC105243748 52.9669 9.08325 0.597624 15.1989 3.59432e-52 1.13371e-48
DESeq2::plotMA(resSex)
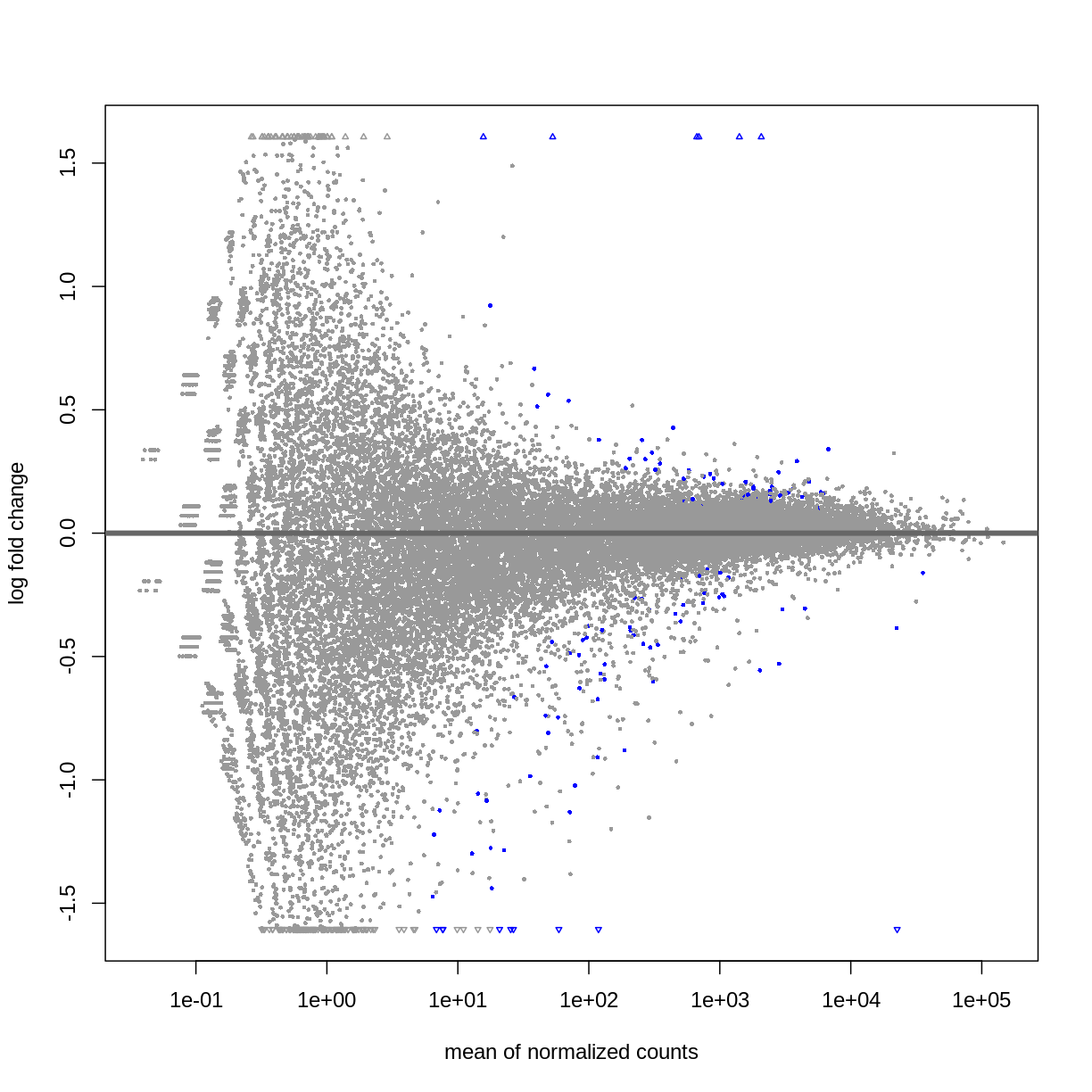
Visualize selected set of genes
Contribute!
Here we intend to practice how to interpret the results from the differential expression analysis. Refer back to the exploratory/QC episode.
vsd <- DESeq2::vst(dds, blind = TRUE)
genes <- rownames(head(resTime[order(resTime$pvalue), ], 10))
heatmapData <- assay(vsd)[genes, ]
heatmapData <- t(scale(t(heatmapData)))
heatmapColAnnot <- data.frame(colData(vsd)[, c("time", "sex")])
idx <- order(vsd$time)
heatmapData <- heatmapData[, idx]
heatmapColAnnot <- HeatmapAnnotation(df = heatmapColAnnot[idx, ])
ComplexHeatmap::Heatmap(heatmapData,
top_annotation = heatmapColAnnot,
cluster_rows = TRUE, cluster_columns = FALSE)
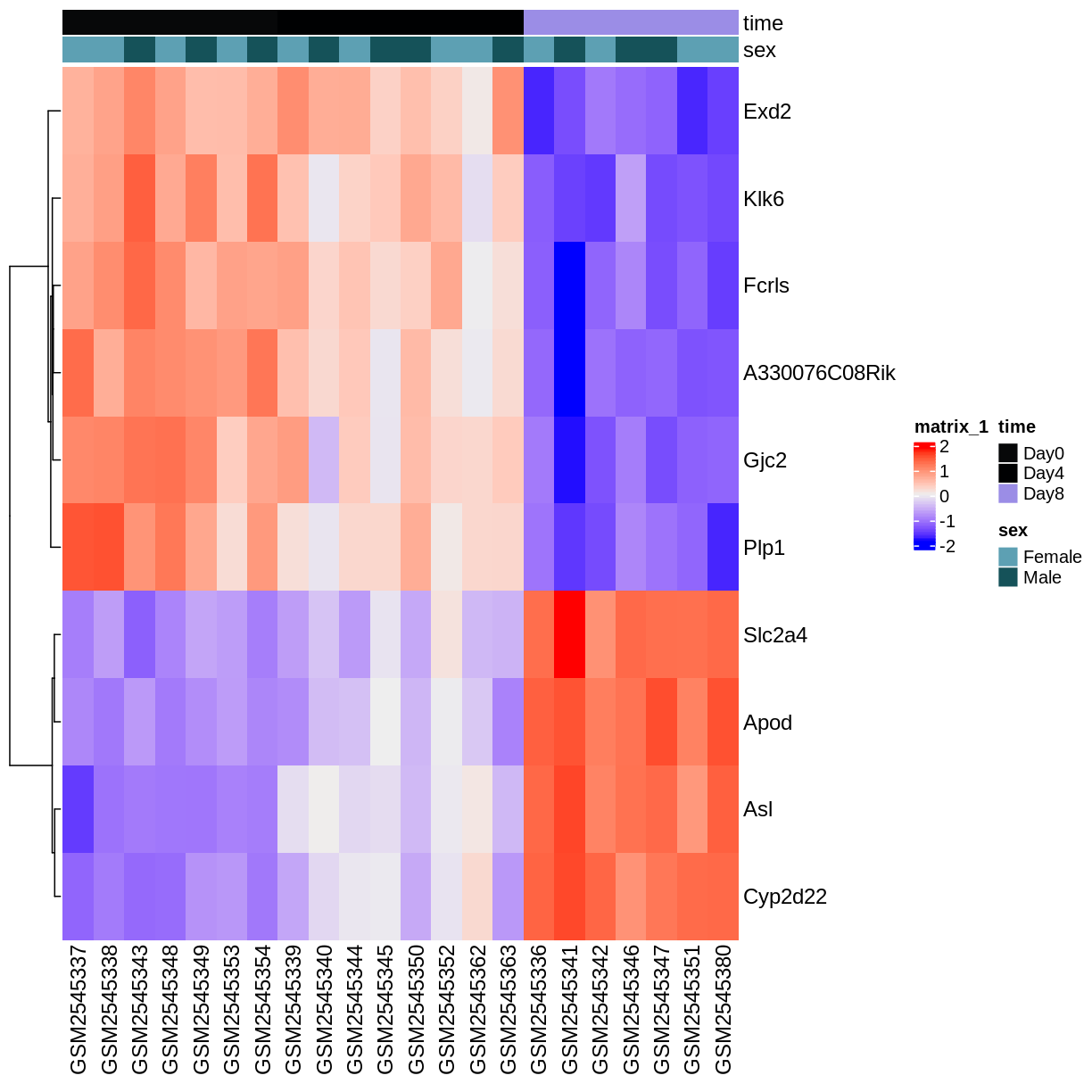
Key Points
Key point 1
Gene set analysis
Overview
Teaching: XX min
Exercises: XX minQuestions
How do we find differentially expressed pathways?
Objectives
Explain how to find differentially expressed pathways with gene set analysis in R.
Understand how differentially expressed genes can enrich a gene set.
Explain how to perform a gene set analysis in R, using clusterProfiler.
Contribute!
This episode is intended to introduce the concept of how to carry out a functional analysis of a subset of differentially expressed (DE) genes, by means of assessing how significantly DE genes enrich gene sets of our interest.
First, we are going to explore the basic concept of enriching a gene set with differentially expressed (DE) genes. Recall the differential expression analysis.
library(SummarizedExperiment)
library(DESeq2)
se <- readRDS("data/GSE96870_se.rds")
dds <- DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"],
design = ~ sex + time)
Warning in DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"], design = ~sex + : some variables in design formula are
characters, converting to factors
dds <- DESeq2::DESeq(dds)
Fetch results for the contrast between male and female mice.
resSex <- DESeq2::results(dds, contrast = c("sex", "Male", "Female"))
summary(resSex)
out of 32652 with nonzero total read count
adjusted p-value < 0.1
LFC > 0 (up) : 53, 0.16%
LFC < 0 (down) : 71, 0.22%
outliers [1] : 10, 0.031%
low counts [2] : 13717, 42%
(mean count < 6)
[1] see 'cooksCutoff' argument of ?results
[2] see 'independentFiltering' argument of ?results
Select DE genes between males and females with FDR < 5%.
sexDE <- as.data.frame(subset(resSex, padj < 0.05))
dim(sexDE)
[1] 54 6
sexDE <- sexDE[order(abs(sexDE$log2FoldChange), decreasing=TRUE), ]
head(sexDE)
baseMean log2FoldChange lfcSE stat pvalue padj
Eif2s3y 1410.8750 12.62514 0.5652155 22.33685 1.620659e-110 1.022366e-106
Kdm5d 692.1672 12.55386 0.5936267 21.14773 2.895664e-99 1.370011e-95
Uty 667.4375 12.01728 0.5935911 20.24505 3.927797e-91 1.486671e-87
Ddx3y 2072.9436 11.87241 0.3974927 29.86825 5.087220e-196 4.813782e-192
Xist 22603.0359 -11.60429 0.3362822 -34.50761 6.168523e-261 1.167393e-256
LOC105243748 52.9669 9.08325 0.5976242 15.19893 3.594320e-52 1.133708e-48
sexDEgenes <- rownames(sexDE)
head(sexDEgenes)
[1] "Eif2s3y" "Kdm5d" "Uty" "Ddx3y" "Xist" "LOC105243748"
length(sexDEgenes)
[1] 54
Enrichment of a curated gene set
Contribute!
Here we illustrate how to assess the enrichment of one gene set we curate ourselves with our subset of DE genes with sex-specific expression. Here we form such a gene set with genes from sex chromosomes. Could you think of another more accurate gene set formed by genes with sex-specific expression?
Build a gene set formed by genes located in the sex chromosomes X and Y.
xygenes <- rownames(se)[decode(seqnames(rowRanges(se)) %in% c("X", "Y"))]
length(xygenes)
[1] 2123
Build a contingency table and conduct a one-tailed Fisher’s exact test that verifies the association between genes being DE between male and female mice and being located in a sex chromosome.
N <- nrow(se)
n <- length(sexDEgenes)
m <- length(xygenes)
k <- length(intersect(xygenes, sexDEgenes))
dnames <- list(GS=c("inside", "outside"), DE=c("yes", "no"))
t <- matrix(c(k, n-k, m-k, N+k-n-m),
nrow=2, ncol=2, dimnames=dnames)
t
DE
GS yes no
inside 18 2105
outside 36 39627
fisher.test(t, alternative="greater")
Fisher's Exact Test for Count Data
data: t
p-value = 7.944e-11
alternative hypothesis: true odds ratio is greater than 1
95 percent confidence interval:
5.541517 Inf
sample estimates:
odds ratio
9.411737
Gene ontology analysis with clusterProfiler
Contribute!
Here we illustrate how to assess the enrichment on the entire collection of Gene Ontology (GO) gene sets using the package clusterProfiler. Could we illustrate any missing important feature of this package for this analysis objective? Could we briefly mention other packages that may be useful for this task?
Second, let’s perform a gene set analysis for an entire collection of gene sets using the Bioconductor package clusterProfiler. For this purpose, we will fetch the results for the contrast between two time points.
resTime <- DESeq2::results(dds, contrast = c("time", "Day8", "Day0"))
summary(resTime)
out of 32652 with nonzero total read count
adjusted p-value < 0.1
LFC > 0 (up) : 4472, 14%
LFC < 0 (down) : 4276, 13%
outliers [1] : 10, 0.031%
low counts [2] : 8732, 27%
(mean count < 1)
[1] see 'cooksCutoff' argument of ?results
[2] see 'independentFiltering' argument of ?results
Select DE genes between Day0 and Day8 with FDR < 5% and minimum 1.5-fold
change.
timeDE <- as.data.frame(subset(resTime, padj < 0.05 & abs(log2FoldChange) > log2(1.5)))
dim(timeDE)
[1] 2110 6
timeDE <- timeDE[order(abs(timeDE$log2FoldChange), decreasing=TRUE), ]
head(timeDE)
baseMean log2FoldChange lfcSE stat pvalue padj
LOC105245444 2.441873 4.768938 0.9013067 5.291138 1.215573e-07 1.800765e-06
LOC105246405 9.728219 4.601505 0.6101832 7.541186 4.657174e-14 2.507951e-12
4933427D06Rik 1.480365 4.556126 1.0318402 4.415535 1.007607e-05 9.169093e-05
A930006I01Rik 2.312732 -4.353155 0.9176026 -4.744053 2.094837e-06 2.252139e-05
LOC105245223 3.272536 4.337202 0.8611255 5.036666 4.737099e-07 6.047199e-06
A530053G22Rik 1.554735 4.243903 1.0248977 4.140806 3.460875e-05 2.720142e-04
timeDEgenes <- rownames(timeDE)
head(timeDEgenes)
[1] "LOC105245444" "LOC105246405" "4933427D06Rik" "A930006I01Rik" "LOC105245223" "A530053G22Rik"
length(timeDEgenes)
[1] 2110
Call the enrichGO() function from
clusterProfiler
as follows.
library(clusterProfiler)
library(org.Mm.eg.db)
library(enrichplot)
resTimeGO <- enrichGO(gene = timeDEgenes,
keyType = "SYMBOL",
universe = rownames(se),
OrgDb = org.Mm.eg.db,
ont = "BP",
pvalueCutoff = 0.01,
qvalueCutoff = 0.01)
dim(resTimeGO)
[1] 32 9
head(resTimeGO)
ID Description GeneRatio BgRatio pvalue
GO:0071674 GO:0071674 mononuclear cell migration 32/1142 166/20469 6.296646e-10
GO:0035456 GO:0035456 response to interferon-beta 16/1142 48/20469 3.291098e-09
GO:0050900 GO:0050900 leukocyte migration 49/1142 355/20469 4.355926e-09
GO:0030595 GO:0030595 leukocyte chemotaxis 34/1142 214/20469 3.265849e-08
GO:0035458 GO:0035458 cellular response to interferon-beta 13/1142 38/20469 6.940867e-08
GO:0002523 GO:0002523 leukocyte migration involved in inflammatory response 10/1142 23/20469 1.649290e-07
p.adjust qvalue
GO:0071674 3.080319e-06 2.852712e-06
GO:0035456 7.103063e-06 6.578212e-06
GO:0050900 7.103063e-06 6.578212e-06
GO:0030595 3.994133e-05 3.699003e-05
GO:0035458 6.790944e-05 6.289156e-05
GO:0002523 1.344721e-04 1.245358e-04
geneID
GO:0071674 Tnfsf18/Aire/Ccl17/Ccr7/Nlrp12/Ccl2/Retnlg/Apod/Il12a/Ccl5/Fpr2/Fut7/Ccl7/Spn/Itgb3/Grem1/Ptk2b/Lgals3/Adam8/Dusp1/Ch25h/Nbl1/Alox5/Padi2/Plg/Calr/Ager/Ccl6/Mdk/Itga4/Hsd3b7/Trpm4
GO:0035456 Tgtp1/Tgtp2/F830016B08Rik/Iigp1/Ifitm6/Igtp/Gm4951/Bst2/Irgm1/Gbp6/Ifi47/Aim2/Ifitm7/Irgm2/Ifit1/Ifi204
GO:0050900 Tnfsf18/Aire/Ccl17/Ccr7/Nlrp12/Bst1/Ccl2/Retnlg/Ppbp/Cxcl5/Apod/Il12a/Ccl5/Fpr2/Umodl1/Fut7/Ccl7/Ccl28/Spn/Sell/Itgb3/Grem1/Cxcl1/Ptk2b/Lgals3/Adam8/Pf4/Dusp1/Ch25h/S100a8/Nbl1/Alox5/Padi2/Plg/Edn3/Il33/Ptn/Ada/Calr/Ager/Ccl6/Prex1/Aoc3/Itgam/Mdk/Itga4/Hsd3b7/P2ry12/Trpm4
GO:0030595 Tnfsf18/Ccl17/Ccr7/Bst1/Ccl2/Retnlg/Ppbp/Cxcl5/Il12a/Ccl5/Fpr2/Ccl7/Sell/Grem1/Cxcl1/Ptk2b/Lgals3/Adam8/Pf4/Dusp1/Ch25h/S100a8/Nbl1/Alox5/Padi2/Edn3/Ptn/Calr/Ccl6/Prex1/Itgam/Mdk/Hsd3b7/Trpm4
GO:0035458 Tgtp1/Tgtp2/F830016B08Rik/Iigp1/Igtp/Gm4951/Irgm1/Gbp6/Ifi47/Aim2/Irgm2/Ifit1/Ifi204
GO:0002523 Ccl2/Ppbp/Fut7/Adam8/S100a8/Alox5/Ptn/Aoc3/Itgam/Mdk
Count
GO:0071674 32
GO:0035456 16
GO:0050900 49
GO:0030595 34
GO:0035458 13
GO:0002523 10
Let’s build a more readable table of results.
library(kableExtra)
resTimeGOtab <- as.data.frame(resTimeGO)
resTimeGOtab$ID <- NULL
resTimeGOtab$geneID <- sapply(strsplit(resTimeGO$geneID, "/"), paste, collapse=", ")
ktab <- kable(resTimeGOtab, row.names=TRUE, caption="GO results for DE genes between time points.")
kable_styling(ktab, bootstrap_options=c("stripped", "hover", "responsive"), fixed_thead=TRUE)
| Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|
| GO:0071674 | mononuclear cell migration | 32/1142 | 166/20469 | 0.00e+00 | 0.0000031 | 0.0000029 | Tnfsf18, Aire, Ccl17, Ccr7, Nlrp12, Ccl2, Retnlg, Apod, Il12a, Ccl5, Fpr2, Fut7, Ccl7, Spn, Itgb3, Grem1, Ptk2b, Lgals3, Adam8, Dusp1, Ch25h, Nbl1, Alox5, Padi2, Plg, Calr, Ager, Ccl6, Mdk, Itga4, Hsd3b7, Trpm4 | 32 |
| GO:0035456 | response to interferon-beta | 16/1142 | 48/20469 | 0.00e+00 | 0.0000071 | 0.0000066 | Tgtp1, Tgtp2, F830016B08Rik, Iigp1, Ifitm6, Igtp, Gm4951, Bst2, Irgm1, Gbp6, Ifi47, Aim2, Ifitm7, Irgm2, Ifit1, Ifi204 | 16 |
| GO:0050900 | leukocyte migration | 49/1142 | 355/20469 | 0.00e+00 | 0.0000071 | 0.0000066 | Tnfsf18, Aire, Ccl17, Ccr7, Nlrp12, Bst1, Ccl2, Retnlg, Ppbp, Cxcl5, Apod, Il12a, Ccl5, Fpr2, Umodl1, Fut7, Ccl7, Ccl28, Spn, Sell, Itgb3, Grem1, Cxcl1, Ptk2b, Lgals3, Adam8, Pf4, Dusp1, Ch25h, S100a8, Nbl1, Alox5, Padi2, Plg, Edn3, Il33, Ptn, Ada, Calr, Ager, Ccl6, Prex1, Aoc3, Itgam, Mdk, Itga4, Hsd3b7, P2ry12, Trpm4 | 49 |
| GO:0030595 | leukocyte chemotaxis | 34/1142 | 214/20469 | 0.00e+00 | 0.0000399 | 0.0000370 | Tnfsf18, Ccl17, Ccr7, Bst1, Ccl2, Retnlg, Ppbp, Cxcl5, Il12a, Ccl5, Fpr2, Ccl7, Sell, Grem1, Cxcl1, Ptk2b, Lgals3, Adam8, Pf4, Dusp1, Ch25h, S100a8, Nbl1, Alox5, Padi2, Edn3, Ptn, Calr, Ccl6, Prex1, Itgam, Mdk, Hsd3b7, Trpm4 | 34 |
| GO:0035458 | cellular response to interferon-beta | 13/1142 | 38/20469 | 1.00e-07 | 0.0000679 | 0.0000629 | Tgtp1, Tgtp2, F830016B08Rik, Iigp1, Igtp, Gm4951, Irgm1, Gbp6, Ifi47, Aim2, Irgm2, Ifit1, Ifi204 | 13 |
| GO:0002523 | leukocyte migration involved in inflammatory response | 10/1142 | 23/20469 | 2.00e-07 | 0.0001345 | 0.0001245 | Ccl2, Ppbp, Fut7, Adam8, S100a8, Alox5, Ptn, Aoc3, Itgam, Mdk | 10 |
| GO:0071675 | regulation of mononuclear cell migration | 21/1142 | 107/20469 | 4.00e-07 | 0.0002770 | 0.0002565 | Tnfsf18, Aire, Ccr7, Ccl2, Apod, Il12a, Ccl5, Fpr2, Spn, Itgb3, Grem1, Ptk2b, Lgals3, Adam8, Dusp1, Nbl1, Padi2, Calr, Ager, Mdk, Itga4 | 21 |
| GO:0002685 | regulation of leukocyte migration | 31/1142 | 213/20469 | 9.00e-07 | 0.0005212 | 0.0004827 | Tnfsf18, Aire, Ccr7, Bst1, Ccl2, Apod, Il12a, Ccl5, Fpr2, Fut7, Ccl28, Spn, Sell, Itgb3, Grem1, Ptk2b, Lgals3, Adam8, Dusp1, Nbl1, Padi2, Edn3, Il33, Ptn, Ada, Calr, Ager, Aoc3, Mdk, Itga4, P2ry12 | 31 |
| GO:0050953 | sensory perception of light stimulus | 26/1142 | 161/20469 | 1.00e-06 | 0.0005212 | 0.0004827 | Aipl1, Vsx2, Nxnl2, Lrit3, Cryba2, Bfsp2, Lrat, Gabrr2, Lum, Rlbp1, Pde6g, Gpr179, Col1a1, Cplx3, Best1, Ush1g, Rs1, Rdh5, Guca1b, Th, Ppef2, Rbp4, Olfm2, Rom1, Vsx1, Rpe65 | 26 |
| GO:0060326 | cell chemotaxis | 38/1142 | 298/20469 | 1.70e-06 | 0.0008479 | 0.0007853 | Tnfsf18, Ccl17, Ccr7, Bst1, Ccl2, Retnlg, Ppbp, Cxcl5, Nr4a1, Il12a, Ccl5, Fpr2, Ccl7, Ccl28, Sell, Grem1, Cxcl1, Ptk2b, Lgals3, Adam8, Pf4, Dusp1, Ch25h, S100a8, Nbl1, Alox5, Padi2, Edn3, Ptn, Plxnb3, Calr, Lpar1, Ccl6, Prex1, Itgam, Mdk, Hsd3b7, Trpm4 | 38 |
| GO:0007601 | visual perception | 25/1142 | 157/20469 | 2.00e-06 | 0.0008935 | 0.0008275 | Aipl1, Vsx2, Nxnl2, Lrit3, Cryba2, Bfsp2, Lrat, Gabrr2, Lum, Rlbp1, Pde6g, Gpr179, Col1a1, Cplx3, Best1, Rs1, Rdh5, Guca1b, Th, Ppef2, Rbp4, Olfm2, Rom1, Vsx1, Rpe65 | 25 |
| GO:0097529 | myeloid leukocyte migration | 30/1142 | 214/20469 | 3.10e-06 | 0.0012605 | 0.0011674 | Tnfsf18, Ccl17, Ccr7, Bst1, Ccl2, Retnlg, Ppbp, Cxcl5, Ccl5, Fpr2, Umodl1, Fut7, Ccl7, Sell, Grem1, Cxcl1, Ptk2b, Lgals3, Adam8, Pf4, Dusp1, S100a8, Nbl1, Edn3, Ager, Ccl6, Prex1, Itgam, Mdk, P2ry12 | 30 |
| GO:1990266 | neutrophil migration | 21/1142 | 122/20469 | 3.70e-06 | 0.0013884 | 0.0012858 | Ccl17, Ccr7, Bst1, Ccl2, Ppbp, Cxcl5, Ccl5, Umodl1, Fut7, Ccl7, Sell, Cxcl1, Lgals3, Adam8, Pf4, S100a8, Edn3, Ccl6, Prex1, Itgam, Mdk | 21 |
| GO:0030593 | neutrophil chemotaxis | 18/1142 | 98/20469 | 7.10e-06 | 0.0024639 | 0.0022819 | Ccl17, Ccr7, Bst1, Ccl2, Ppbp, Cxcl5, Ccl5, Ccl7, Sell, Cxcl1, Lgals3, Pf4, S100a8, Edn3, Ccl6, Prex1, Itgam, Mdk | 18 |
| GO:0071677 | positive regulation of mononuclear cell migration | 14/1142 | 64/20469 | 9.10e-06 | 0.0029614 | 0.0027426 | Tnfsf18, Ccr7, Ccl2, Il12a, Ccl5, Fpr2, Spn, Itgb3, Ptk2b, Lgals3, Adam8, Calr, Ager, Itga4 | 14 |
| GO:0030198 | extracellular matrix organization | 36/1142 | 297/20469 | 1.02e-05 | 0.0029937 | 0.0027725 | Nepn, Has2, Fbln5, Adamts14, Nox1, Adamtsl2, Mmp8, Lum, Itgb3, Nid1, Grem1, Elf3, Col5a3, Lgals3, Col1a1, Serpinh1, Col27a1, Loxl4, Agt, Kazald1, Colq, Pxdn, Plg, Col11a2, Col15a1, P4ha1, Mpzl3, Mmp15, Has3, Cav1, Ccdc80, Spint1, Abi3bp, Adamts16, Col14a1, Cyp1b1 | 36 |
| GO:0043062 | extracellular structure organization | 36/1142 | 298/20469 | 1.10e-05 | 0.0029937 | 0.0027725 | Nepn, Has2, Fbln5, Adamts14, Nox1, Adamtsl2, Mmp8, Lum, Itgb3, Nid1, Grem1, Elf3, Col5a3, Lgals3, Col1a1, Serpinh1, Col27a1, Loxl4, Agt, Kazald1, Colq, Pxdn, Plg, Col11a2, Col15a1, P4ha1, Mpzl3, Mmp15, Has3, Cav1, Ccdc80, Spint1, Abi3bp, Adamts16, Col14a1, Cyp1b1 | 36 |
| GO:0045229 | external encapsulating structure organization | 36/1142 | 298/20469 | 1.10e-05 | 0.0029937 | 0.0027725 | Nepn, Has2, Fbln5, Adamts14, Nox1, Adamtsl2, Mmp8, Lum, Itgb3, Nid1, Grem1, Elf3, Col5a3, Lgals3, Col1a1, Serpinh1, Col27a1, Loxl4, Agt, Kazald1, Colq, Pxdn, Plg, Col11a2, Col15a1, P4ha1, Mpzl3, Mmp15, Has3, Cav1, Ccdc80, Spint1, Abi3bp, Adamts16, Col14a1, Cyp1b1 | 36 |
| GO:0034341 | response to interferon-gamma | 21/1142 | 133/20469 | 1.48e-05 | 0.0038065 | 0.0035252 | Ccl17, Gbp4, Ccl2, Tgtp1, H2-Q7, Il12rb1, Ifitm6, Ccl5, Ccl7, Nos2, Nlrc5, Bst2, Irgm1, Gbp6, Capg, Ifitm7, Gbp9, Gbp5, Irgm2, Ccl6, Aqp4 | 21 |
| GO:0036336 | dendritic cell migration | 8/1142 | 23/20469 | 2.12e-05 | 0.0051736 | 0.0047913 | Tnfsf18, Ccr7, Nlrp12, Retnlg, Il12a, Alox5, Calr, Trpm4 | 8 |
| GO:0002819 | regulation of adaptive immune response | 27/1142 | 203/20469 | 2.49e-05 | 0.0058002 | 0.0053716 | Tnfsf18, Ccr7, H2-Q6, H2-Q7, Alox15, Il12a, Il12rb1, H2-Q4, Fut7, Spn, H2-Q2, Irf7, Cd274, Tnfrsf13c, Il33, H2-Q1, Ada, C3, Tfrc, H2-K1, Ager, H2-T23, Tap2, Tnfsf13b, Pla2g4a, Trpm4, Parp3 | 27 |
| GO:0001906 | cell killing | 26/1142 | 193/20469 | 2.79e-05 | 0.0060788 | 0.0056296 | Ccl17, Gzmb, Ccl2, Cxcl5, H2-Q6, H2-Q7, Il12a, H2-Q4, Il18rap, Ccl28, Nos2, Cxcl1, H2-Q2, Lgals3, Pf4, Tap1, H2-Q1, Emp2, Scnn1b, C3, H2-K1, Ager, H2-T23, Itgam, Tap2, Clec7a | 26 |
| GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 28/1142 | 216/20469 | 2.86e-05 | 0.0060788 | 0.0056296 | Ccr7, Nr4a3, Has2, Ccl2, Il12a, Il12rb1, Ccl5, Fut7, Spn, Cd5, Icosl, Il2rg, Adam8, Cd274, Tnfrsf13c, Hsph1, Alox5, Btn2a2, Ada, Tfrc, Cd46, Ager, Xbp1, H2-T23, Tnfsf13b, Mdk, Itga4, Cav1 | 28 |
| GO:1901615 | organic hydroxy compound metabolic process | 50/1142 | 492/20469 | 3.17e-05 | 0.0064643 | 0.0059866 | Hao1, Dio3, Cyp1a1, Nr4a2, Sult1a1, Cyp4f18, Lrat, Ddc, Apoc1, Alox15, Lhcgr, Drd4, Cyp4f15, Acer2, Lcat, Hsd17b1, Ptk2b, Actn3, Akr1c14, Ch25h, Cyp11a1, Dct, Cyp2d22, Dkkl1, Cyp51, Srebf1, Scnn1b, Npc1l1, Moxd1, Msmo1, Hmgcs1, Ctsk, Ebp, Th, Qdpr, Abca1, Itgam, Kcnj6, Pla2g4a, Pmel, Cyb5r2, Cdh3, Rbp4, Hsd3b7, Idh2, Slc5a5, Tg, Rpe65, Gpr37, Cyp1b1 | 50 |
| GO:0097530 | granulocyte migration | 22/1142 | 151/20469 | 3.39e-05 | 0.0066403 | 0.0061496 | Tnfsf18, Ccl17, Ccr7, Bst1, Ccl2, Ppbp, Cxcl5, Ccl5, Umodl1, Fut7, Ccl7, Sell, Cxcl1, Lgals3, Adam8, Pf4, S100a8, Edn3, Ccl6, Prex1, Itgam, Mdk | 22 |
| GO:0007159 | leukocyte cell-cell adhesion | 38/1142 | 340/20469 | 3.71e-05 | 0.0069751 | 0.0064597 | Tnfsf18, Ccr7, Nr4a3, Has2, Slfn1, Ccl2, Il12a, Il12rb1, Ccl5, Fut7, Ccl28, Spn, Cd5, Sell, Icosl, Il2rg, Btn1a1, Lgals3, Adam8, Cd274, Tnfrsf13c, Hsph1, S100a8, Btla, Alox5, Btn2a2, Ada, Tfrc, Cd46, Ass1, Ager, Xbp1, H2-T23, Itgam, Tnfsf13b, Mdk, Itga4, Cav1 | 38 |
| GO:1903037 | regulation of leukocyte cell-cell adhesion | 35/1142 | 304/20469 | 4.03e-05 | 0.0072725 | 0.0067351 | Tnfsf18, Ccr7, Nr4a3, Has2, Slfn1, Ccl2, Il12a, Il12rb1, Ccl5, Fut7, Ccl28, Spn, Cd5, Icosl, Il2rg, Btn1a1, Lgals3, Adam8, Cd274, Tnfrsf13c, Hsph1, Btla, Alox5, Btn2a2, Ada, Tfrc, Cd46, Ass1, Ager, Xbp1, H2-T23, Tnfsf13b, Mdk, Itga4, Cav1 | 35 |
| GO:0031349 | positive regulation of defense response | 30/1142 | 244/20469 | 4.16e-05 | 0.0072725 | 0.0067351 | Tnfsf18, Ccr7, Zbp1, Pla2g3, Il12a, Ccl5, Mmp8, Fpr2, Il18rap, Nlrc5, Cxcl1, Adam8, Irf7, Irgm1, S100a8, Pla2g5, Aim2, Srebf1, Il33, C3, Npas2, Gbp5, Irgm2, Ager, Aoc3, H2-T23, Pla2g4a, Mdk, Ace, Ifi204 | 30 |
| GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 25/1142 | 187/20469 | 4.50e-05 | 0.0075878 | 0.0070271 | Tnfsf18, Ccr7, H2-Q6, H2-Q7, Il12a, Il12rb1, H2-Q4, Fut7, Spn, H2-Q2, Cd274, Tnfrsf13c, Il33, H2-Q1, Ada, C3, Tfrc, H2-K1, Ager, H2-T23, Tap2, Tnfsf13b, Pla2g4a, Trpm4, Parp3 | 25 |
| GO:0072676 | lymphocyte migration | 17/1142 | 102/20469 | 4.66e-05 | 0.0075967 | 0.0070354 | Aire, Ccl17, Ccr7, Ccl2, Apod, Ccl5, Fut7, Ccl7, Spn, Itgb3, Ptk2b, Adam8, Ch25h, Padi2, Ccl6, Itga4, Hsd3b7 | 17 |
| GO:0071621 | granulocyte chemotaxis | 19/1142 | 123/20469 | 5.07e-05 | 0.0079992 | 0.0074081 | Tnfsf18, Ccl17, Ccr7, Bst1, Ccl2, Ppbp, Cxcl5, Ccl5, Ccl7, Sell, Cxcl1, Lgals3, Pf4, S100a8, Edn3, Ccl6, Prex1, Itgam, Mdk | 19 |
| GO:0032103 | positive regulation of response to external stimulus | 41/1142 | 387/20469 | 6.37e-05 | 0.0097332 | 0.0090140 | Tnfsf18, Ccr7, Ccl2, Zbp1, Pla2g3, Ntf3, Il12a, Ccl5, Mmp8, Fpr2, Il18rap, Sell, Casr, Nlrc5, Cxcl1, Ptk2b, Adam8, Irf7, Irgm1, S100a8, Pla2g5, Aim2, Plg, Srebf1, Edn3, Il33, Ptn, C3, Calr, Lpar1, Gbp5, Irgm2, Ager, Aoc3, H2-T23, Pla2g4a, Mdk, Ace, Stx3, P2ry12, Ifi204 | 41 |
Key Points
Key point 1