Differential expression analysis
Overview
Teaching: XX min
Exercises: XX minQuestions
How do we find differentially expressed genes?
Objectives
Explain the steps involved in a differential expression analysis.
Explain how to perform these steps in R, using DESeq2.
Contribute!
This episode is intended to introduce the concepts required to perform differential expression analysis with RNA-seq data. Explain concepts like size factors, count modeling (Negative Binomial), dispersion, interpretation of the test output, multiple testing correction.
suppressPackageStartupMessages({
library(SummarizedExperiment)
library(DESeq2)
library(ggplot2)
library(ExploreModelMatrix)
library(cowplot)
library(ComplexHeatmap)
})
Load data
se <- readRDS("data/GSE96870_se.rds")
Create DESeqDataSet
dds <- DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"],
design = ~ sex + time)
Warning in DESeq2::DESeqDataSet(se[, se$tissue == "Cerebellum"], design = ~sex + : some variables in design formula are
characters, converting to factors
Run DESeq()
Contribute!
The concepts may be clearer if the steps of DESeq() are first performed separately, followed by a note that they can be performed in a single step using DESeq().
dds <- DESeq2::DESeq(dds)
estimating size factors
estimating dispersions
gene-wise dispersion estimates
mean-dispersion relationship
final dispersion estimates
fitting model and testing
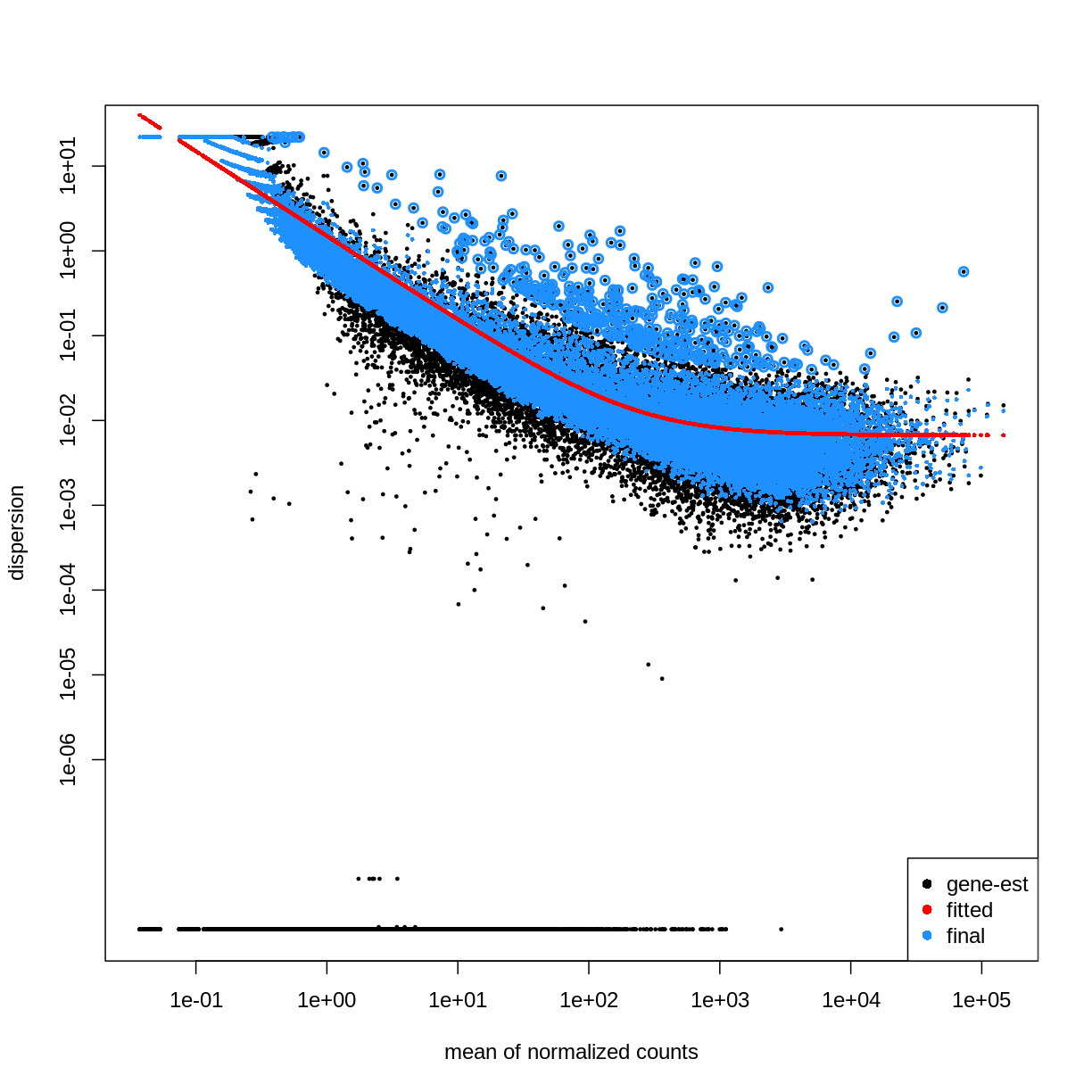
plotDispEsts(dds)
Extract results for specific contrasts
Contribute!
Refer back to the episode about experimental design.
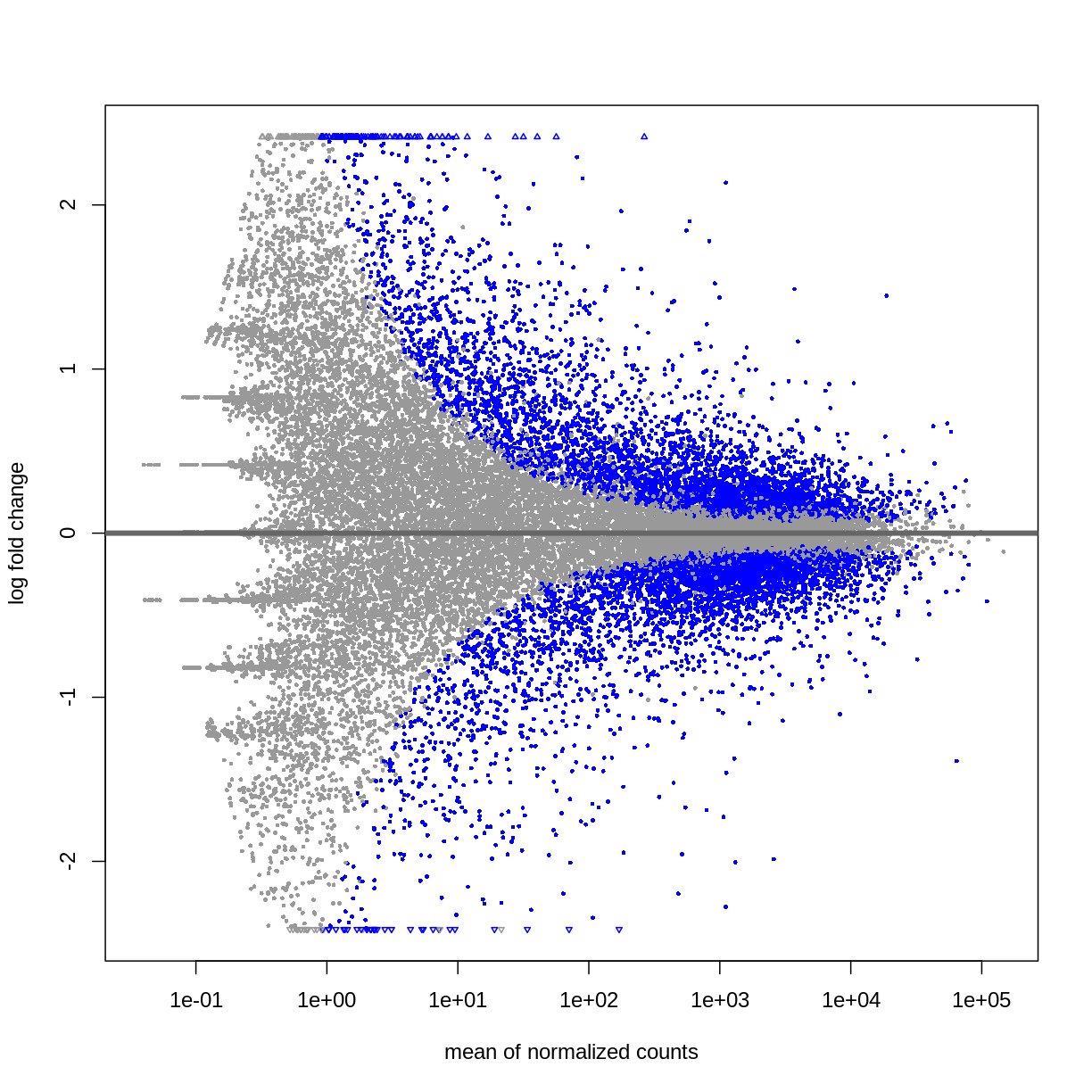
## Day 8 vs Day 0
resTime <- DESeq2::results(dds, contrast = c("time", "Day8", "Day0"))
summary(resTime)
out of 32652 with nonzero total read count
adjusted p-value < 0.1
LFC > 0 (up) : 4472, 14%
LFC < 0 (down) : 4276, 13%
outliers [1] : 10, 0.031%
low counts [2] : 8732, 27%
(mean count < 1)
[1] see 'cooksCutoff' argument of ?results
[2] see 'independentFiltering' argument of ?results
head(resTime[order(resTime$pvalue), ])
log2 fold change (MLE): time Day8 vs Day0
Wald test p-value: time Day8 vs Day0
DataFrame with 6 rows and 6 columns
baseMean log2FoldChange lfcSE stat pvalue padj
<numeric> <numeric> <numeric> <numeric> <numeric> <numeric>
Asl 701.343 1.11733 0.0592541 18.8565 2.59885e-79 6.21386e-75
Apod 18765.146 1.44698 0.0805186 17.9708 3.30147e-72 3.94690e-68
Cyp2d22 2550.480 0.91020 0.0554756 16.4072 1.69794e-60 1.35326e-56
Klk6 546.503 -1.67190 0.1058989 -15.7877 3.78228e-56 2.26086e-52
Fcrls 184.235 -1.94701 0.1279847 -15.2128 2.90708e-52 1.39017e-48
A330076C08Rik 107.250 -1.74995 0.1154279 -15.1606 6.45112e-52 2.57077e-48
DESeq2::plotMA(resTime)
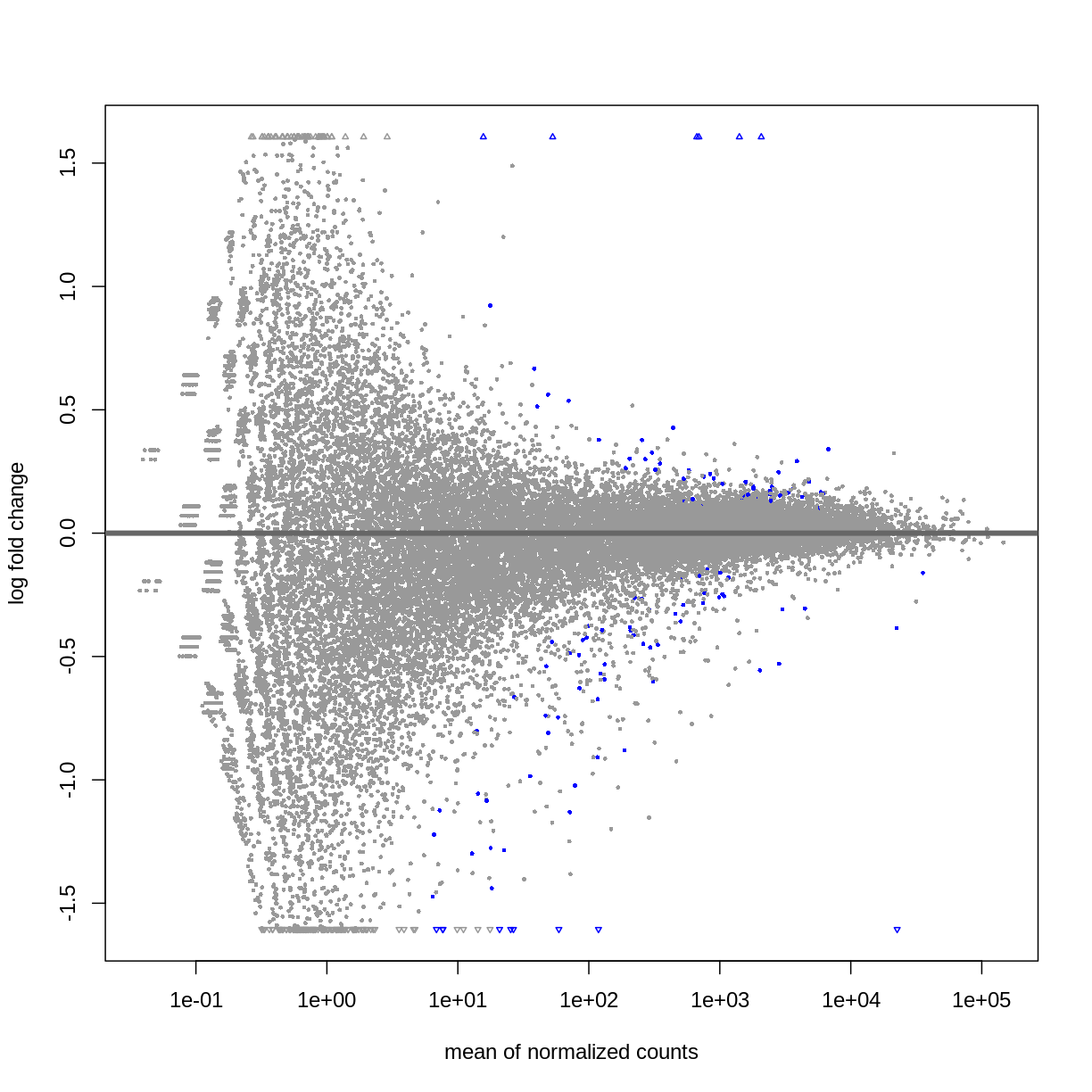
## Male vs Female
resSex <- DESeq2::results(dds, contrast = c("sex", "Male", "Female"))
summary(resSex)
out of 32652 with nonzero total read count
adjusted p-value < 0.1
LFC > 0 (up) : 53, 0.16%
LFC < 0 (down) : 71, 0.22%
outliers [1] : 10, 0.031%
low counts [2] : 13717, 42%
(mean count < 6)
[1] see 'cooksCutoff' argument of ?results
[2] see 'independentFiltering' argument of ?results
head(resSex[order(resSex$pvalue), ])
log2 fold change (MLE): sex Male vs Female
Wald test p-value: sex Male vs Female
DataFrame with 6 rows and 6 columns
baseMean log2FoldChange lfcSE stat pvalue padj
<numeric> <numeric> <numeric> <numeric> <numeric> <numeric>
Xist 22603.0359 -11.60429 0.336282 -34.5076 6.16852e-261 1.16739e-256
Ddx3y 2072.9436 11.87241 0.397493 29.8683 5.08722e-196 4.81378e-192
Eif2s3y 1410.8750 12.62514 0.565216 22.3369 1.62066e-110 1.02237e-106
Kdm5d 692.1672 12.55386 0.593627 21.1477 2.89566e-99 1.37001e-95
Uty 667.4375 12.01728 0.593591 20.2451 3.92780e-91 1.48667e-87
LOC105243748 52.9669 9.08325 0.597624 15.1989 3.59432e-52 1.13371e-48
DESeq2::plotMA(resSex)
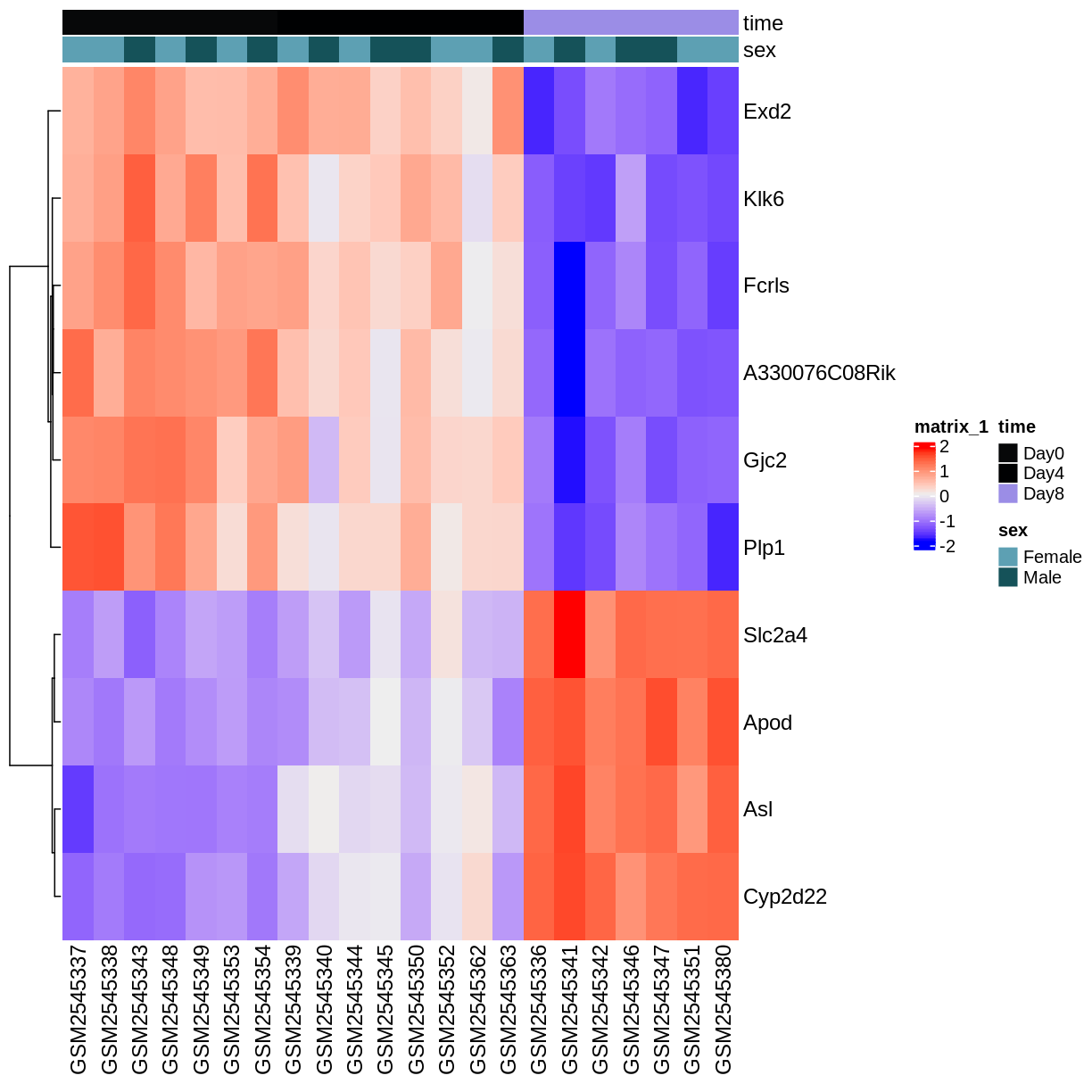
Visualize selected set of genes
Contribute!
Here we intend to practice how to interpret the results from the differential expression analysis. Refer back to the exploratory/QC episode.
vsd <- DESeq2::vst(dds, blind = TRUE)
genes <- rownames(head(resTime[order(resTime$pvalue), ], 10))
heatmapData <- assay(vsd)[genes, ]
heatmapData <- t(scale(t(heatmapData)))
heatmapColAnnot <- data.frame(colData(vsd)[, c("time", "sex")])
idx <- order(vsd$time)
heatmapData <- heatmapData[, idx]
heatmapColAnnot <- HeatmapAnnotation(df = heatmapColAnnot[idx, ])
ComplexHeatmap::Heatmap(heatmapData,
top_annotation = heatmapColAnnot,
cluster_rows = TRUE, cluster_columns = FALSE)
Key Points
Key point 1